Journal Highlights
*The following is a selection of published papers. For specific research papers/research results, please check each laboratory’s website.
2026
-
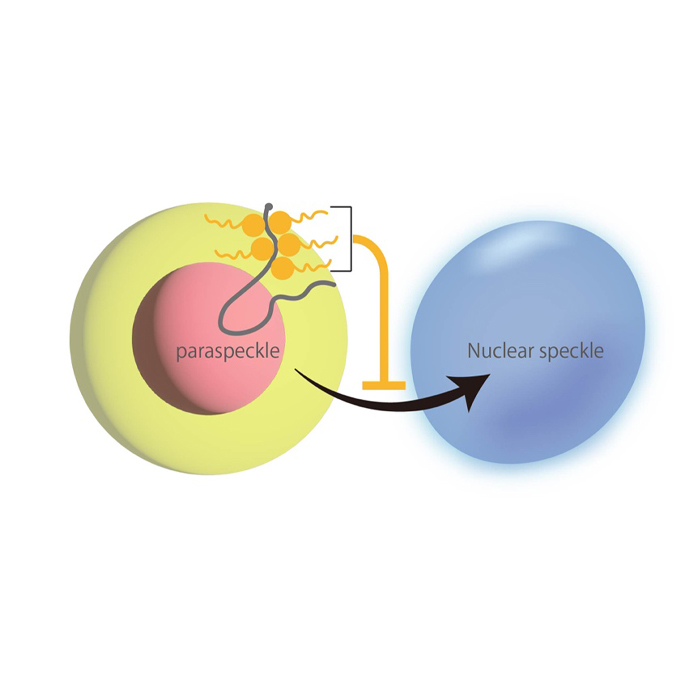
Blocky proline/glutamine patterns in the SFPQ intrinsically disordered region dictate paraspeckle formation as a distinct membraneless organelle
A research team at the University of Osaka has identified a novel amino acid sequence pattern that dictates paraspeckle independence within the nucleoplasmic environment.
RNA Biofunction Laboratory
-
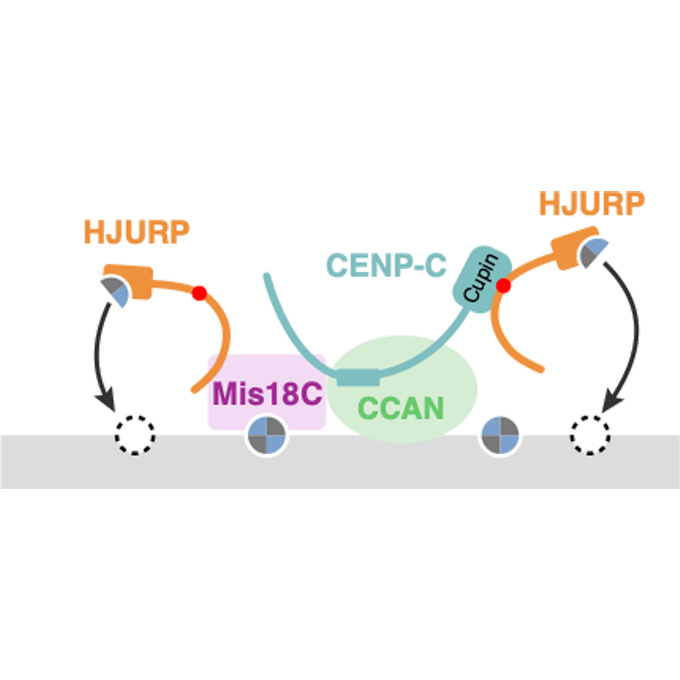
Searching for the centromere: diversity in pathways key for cell division
A research team at The University of Osaka has found a novel pathway for depositing CENP-A into vertebrate centromeres, which is key for microtubule attachment and subsequent cell division
Laboratory of Chromosome Biology
2025
-
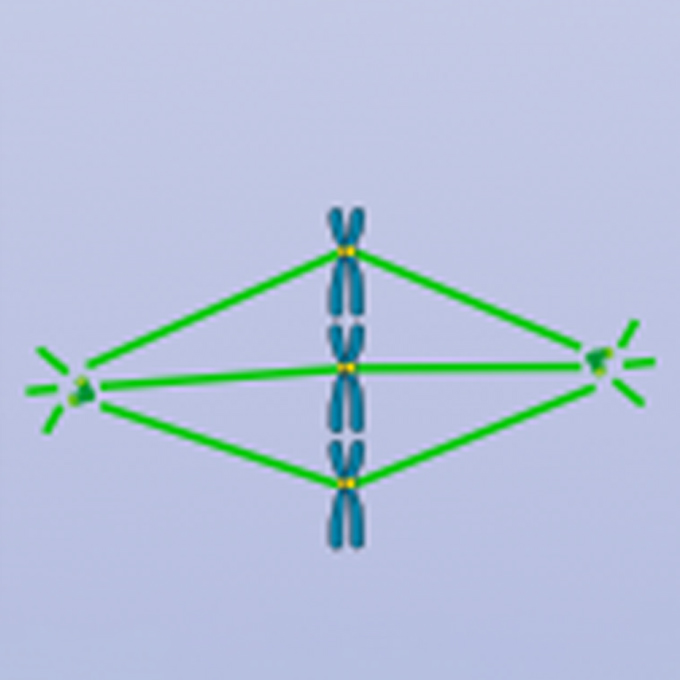
Cooperative motor proteins found to kill cancer cells when dual-inhibited
Discovery reveals how key molecules coordinate chromosome alignment and offers a new anticancer target
Laboratory of Chromosome Biology
-
A β-cap on the FliPQR protein-export channel acts as the cap for initial flagellar rod assembly
-
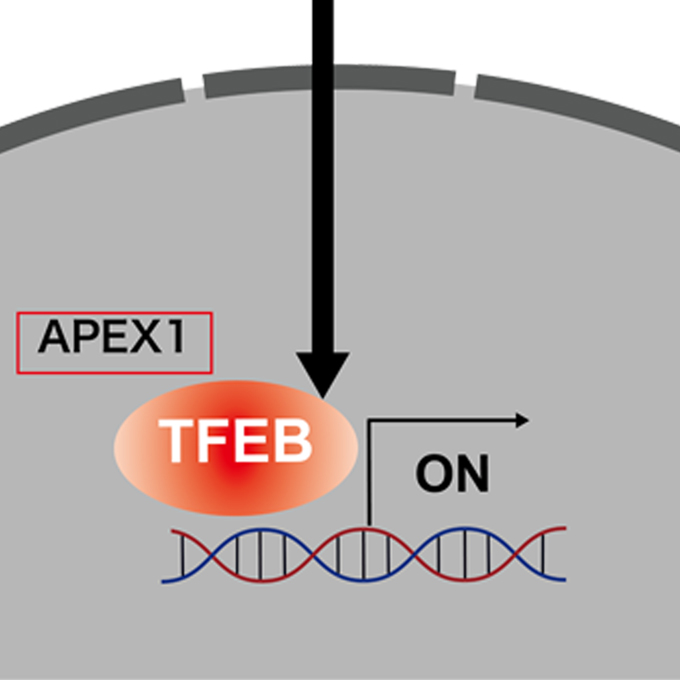
ATG conjugation dependent/independent mechanisms underlie lysosomal stress induced TFEB regulation
-
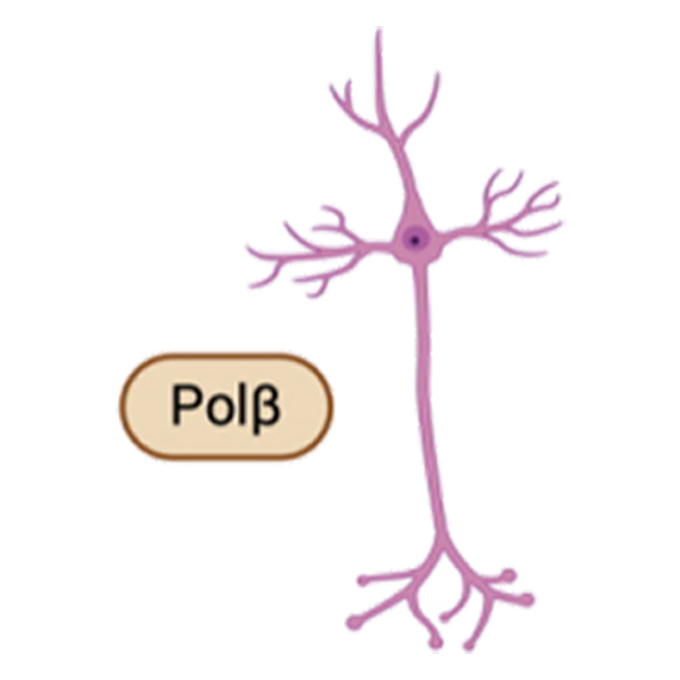
Enzyme protects developing brain from harmful mutations
Loss of Polβ leads to increased mutations in developing neurons, potentially contributing to neurodevelopmental disorders
KOKORO-Biology Group
-
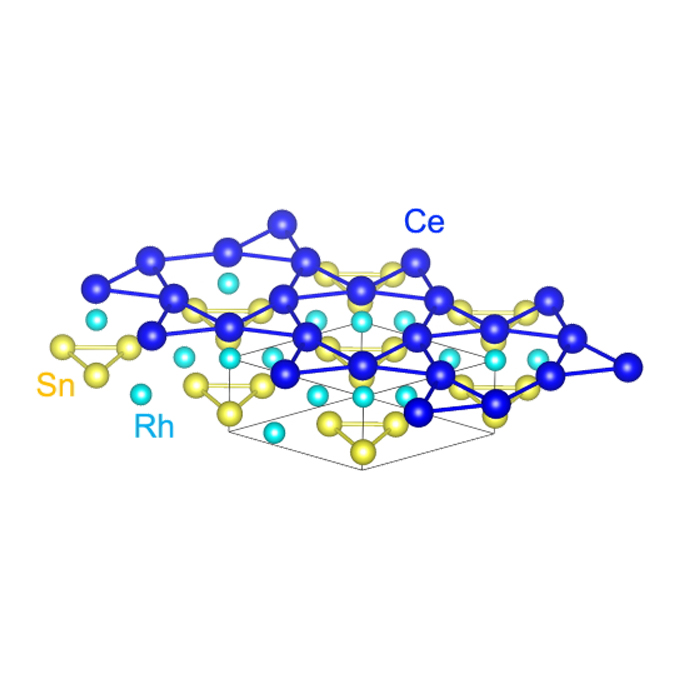
Heavy fermions entangled: Quantum computing's new frontier?
Discovery of Planckian time limit opens doors to novel quantum technologies
Photophysics Laboratory
-
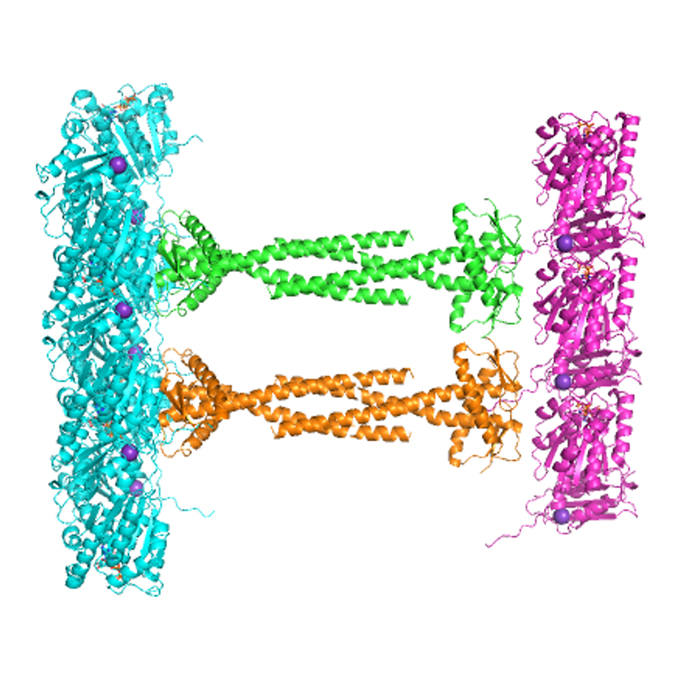
Exploring The Dynamic Partnership Between FtsZ and ZapA Protein
Researchers explore the structure and molecular basis of the interaction between bacterial cell division-associated proteins FtsZ and ZapA
JEOL YOKOGUSHI Research Alliance Laboratories
-
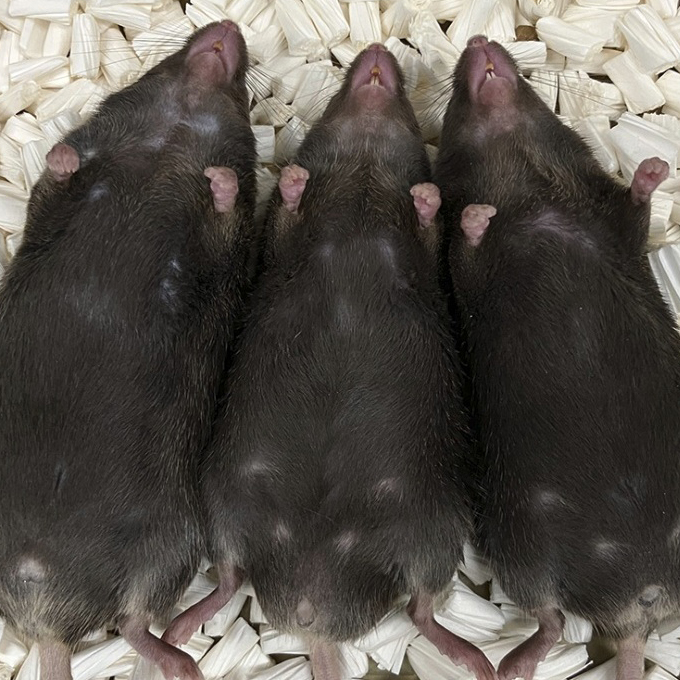
Maternal iron deficiency causes male-to-female sex reversal in mouse embryos
Researchers from The University of Osaka have revealed the crucial role that iron plays in sex determination of mammalian embryos
Laboratory of Epigenome Dynamics
-
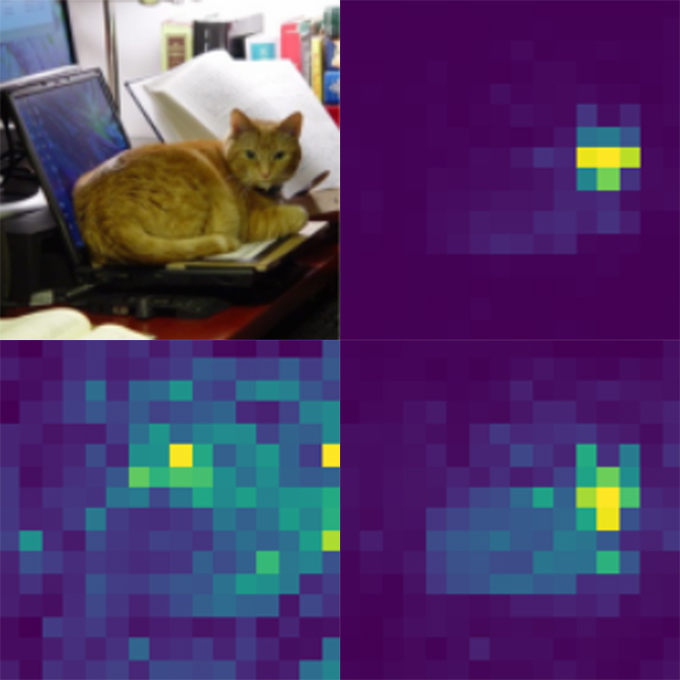
Self-trained vision transformers mimic human gaze with surprising precision
A research team led by The University of Osaka has demonstrated that vision transformers using self-attention mechanisms can spontaneously develop visual attention patterns similar to humans without specific training
Dynamic Brain Network Laboratory
-

Tiny Brain Region Found to Drive Motor Learning in Reaching Movements
Researchers uncover critical role of the parvocellular red nucleus in correcting movement errors, paving the way for future rehabilitation technologies
Dynamic Brain Network Laboratory
-
In silico screening by AlphaFold2 program revealed the potential binding partners of nuage-localizing proteins and piRNA-related proteins
Germline Biology Group
-

Atf3 controls transitioning in female mitochondrial cardiomyopathy as identified by spatial and single-cell transcriptomics
Laboratory of Medical Biochemistry
-

Excitable Ras dynamics-based screens reveal RasGEFX is required for macropinocytosis and random cell migration
Laboratory of Single Molecule Biology
2024
-
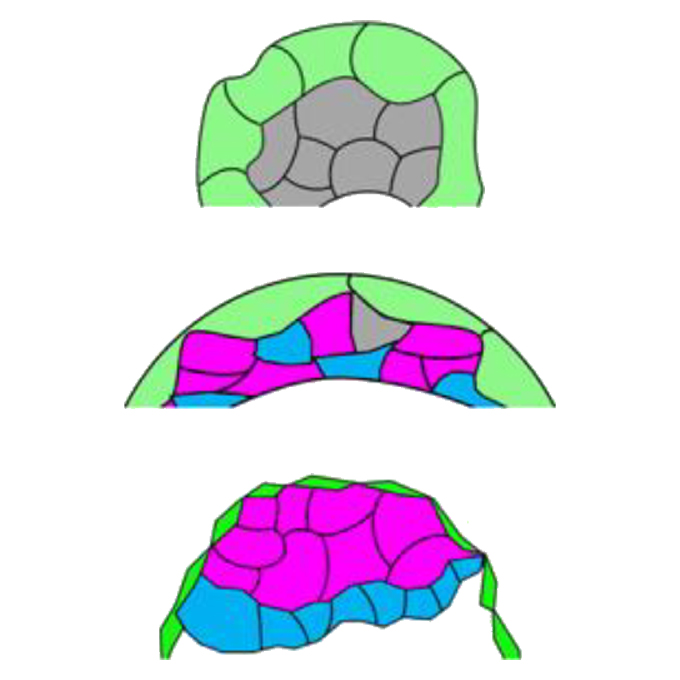
Fate specification triggers a positive feedback loop of TEAD-YAP and NANOG to promote epiblast formation in preimplantation embryos
Laboratory for Embryogenesis
-

High-tech tracking technology streamlines drug discovery
Laboratory of Single Molecule Biology
-
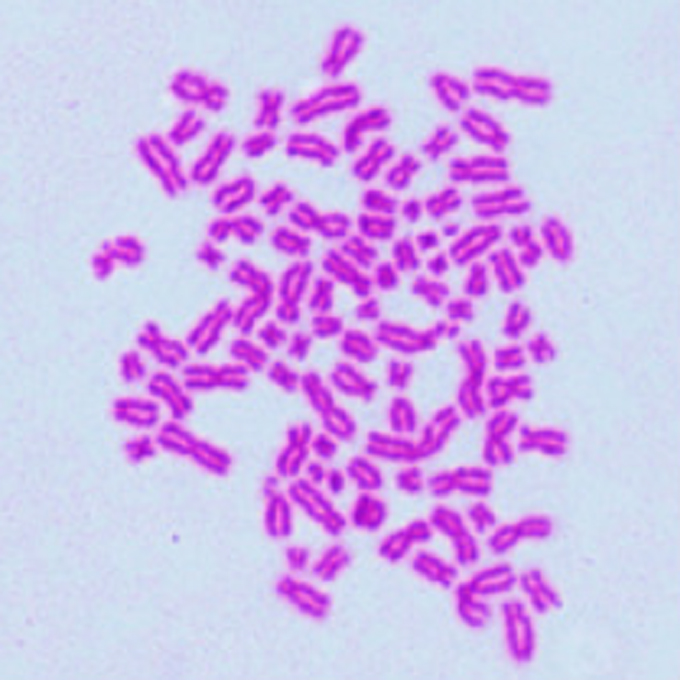
Polyploidy mitigates the impact of DNA damage while simultaneously bearing its burden
Laboratory of Ploidy Pathology
-
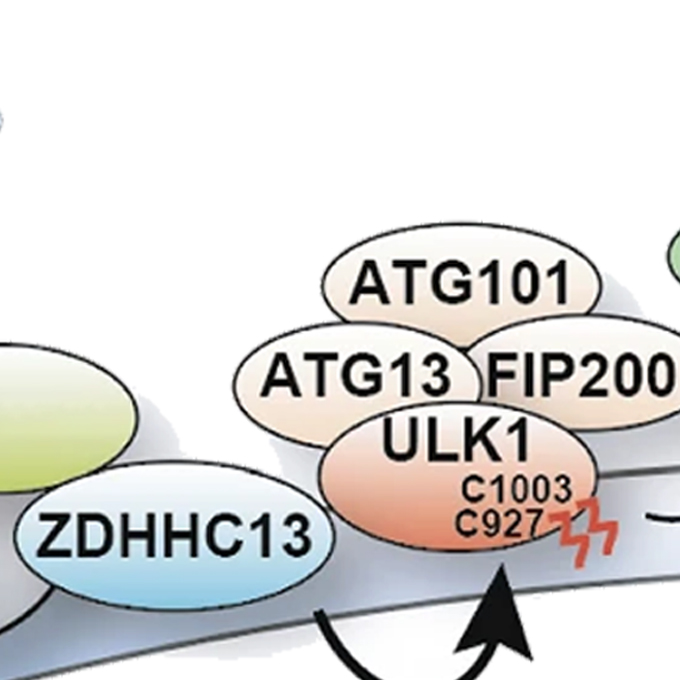
Palmitoylation of ULK1 by ZDHHC13 plays a crucial role in autophagy
-
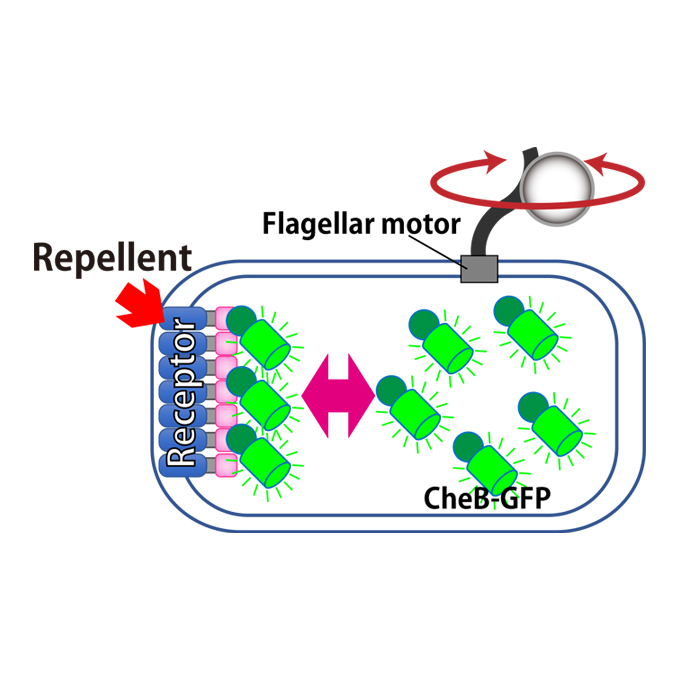
CheB localizes to polar receptor arrays during repellent adaptation
Laboratory of Nano-Biophysics
-
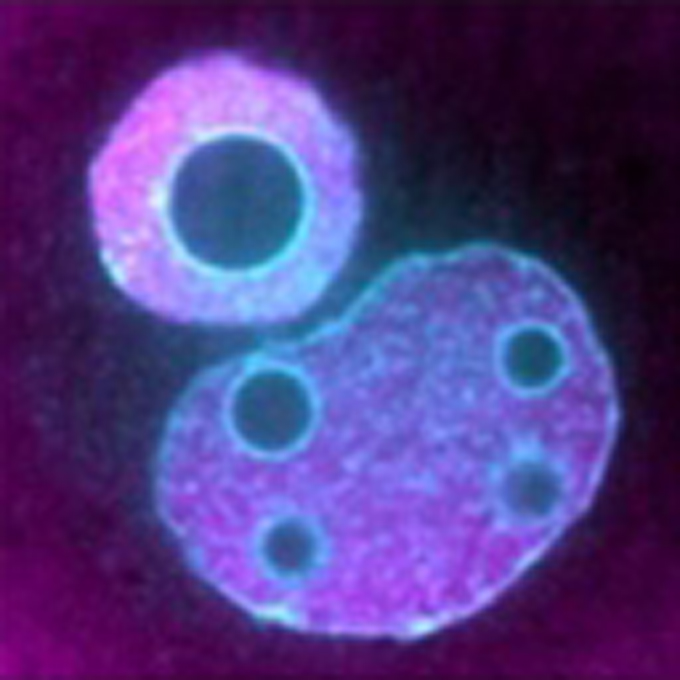
Reconstruction of artificial nuclei with nuclear import activity in living mouse oocytes
-
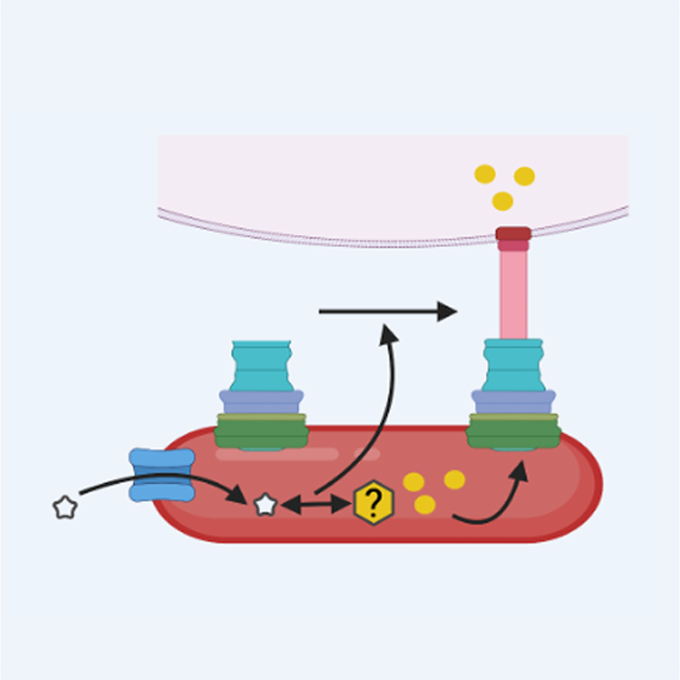
Salmonella Typhimurium exploits host polyamines for assembly of the type 3 secretion machinery
Protonic NanoMachine Group
-
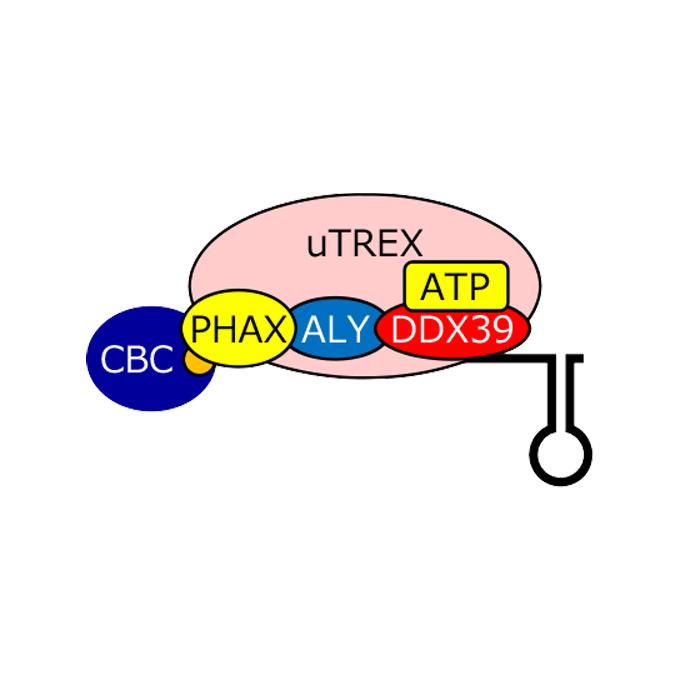
The RNA helicase DDX39 contributes to the nuclear export of spliceosomal U snRNA by loading of PHAX onto RNA
RNA Biofunction Laboratory
-

HemK2 functions for sufficient protein synthesis and RNA stability through eRF1 methylation during Drosophila oogenesis
Germline Biology Group
-
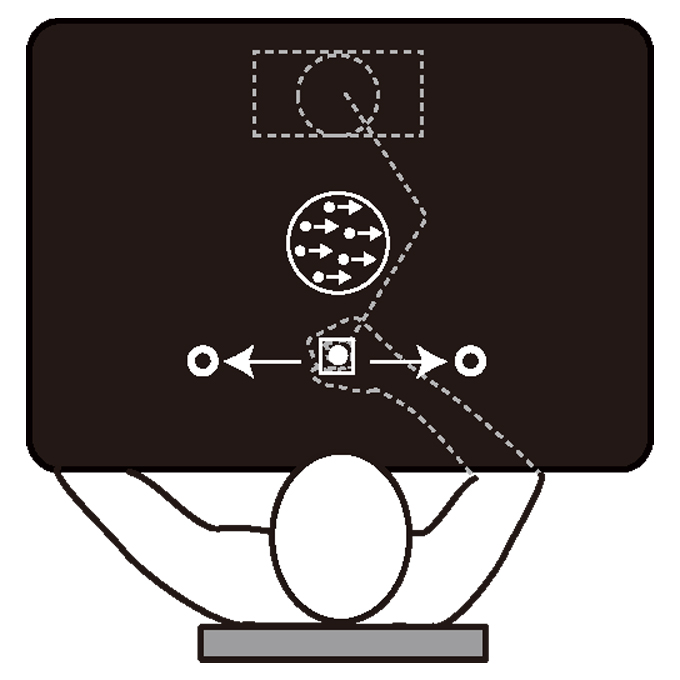
Decision uncertainty as a context for motor memory
-
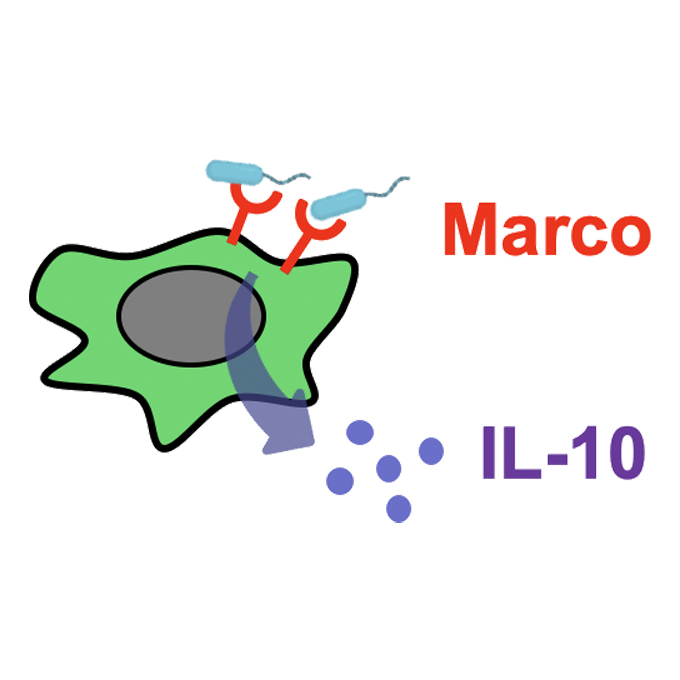
Periportal macrophages protect against commensal-driven liver inflammation
Laboratory of Immunology and Cell Biology
-
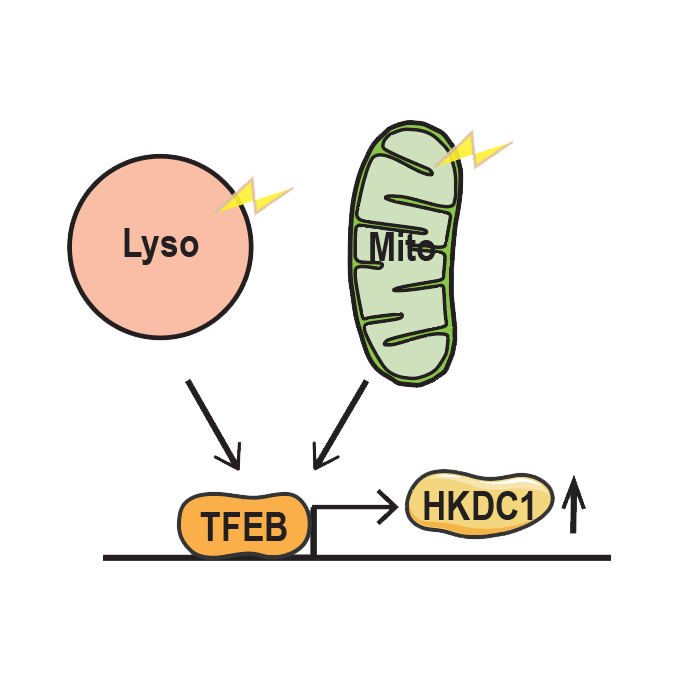
HKDC1, a target of TFEB, is essential to maintain both mitochondrial and lysosomal homeostasis, preventing cellular senescence
Laboratory of Intracellular Membrane Dynamics
2023
-
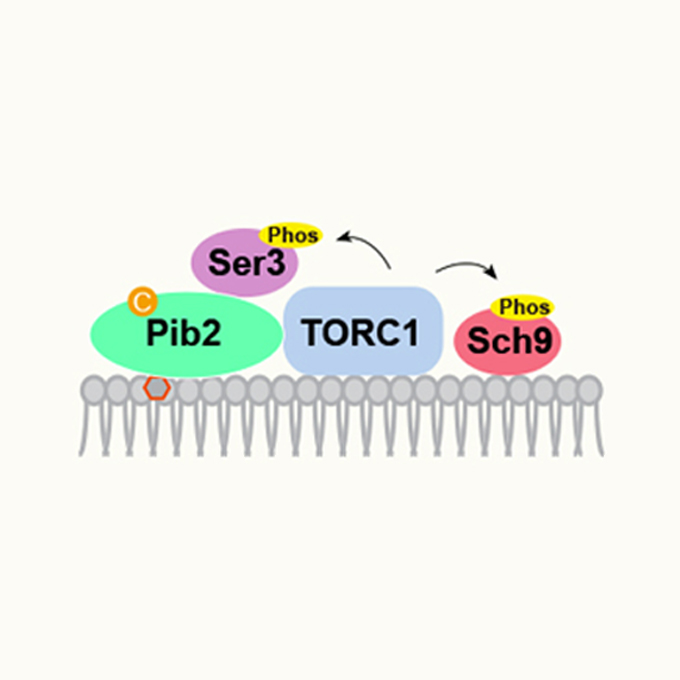
Pib2 is a cysteine sensor involved in TORC1 activation in Saccharomyces cerevisiae
-
Microbes control Drosophila germline stem cell increase and egg maturation through hormonal pathways
Germline Biology Group
-
Repetitive CREB-DNA interactions at gene loci predetermined by CBP induce activity-dependent gene expression in human cortical neurons
-
Microautophagy regulated by STK38 and GABARAPs is essential to repair lysosomes and prevent aging
Laboratory of Intracellular Membrane Dynamics
-
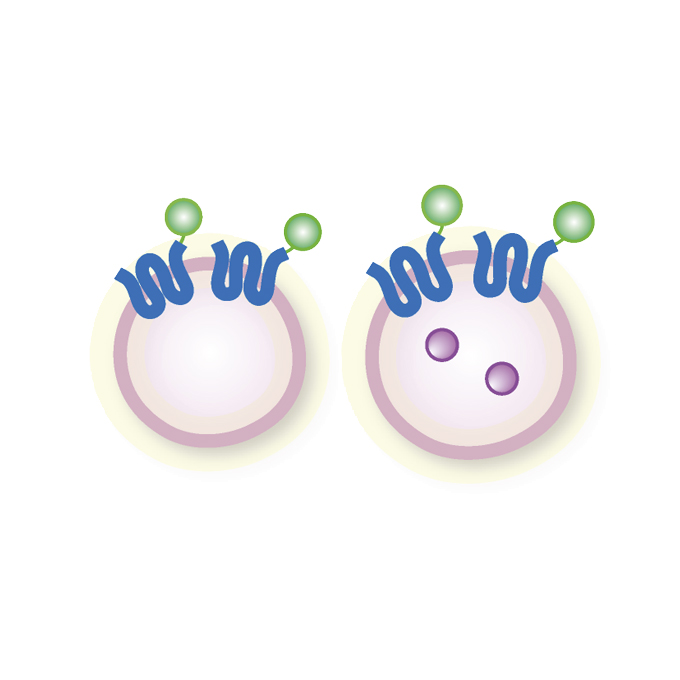
The TMEM192-mKeima probe specifically assays lysophagy and reveals its initial steps
Laboratory of Intracellular Membrane Dynamics
-

Two-dimensional heavy fermion in monoatomic-layer Kondo lattice YbCu2
Photophysics Laboratory
-
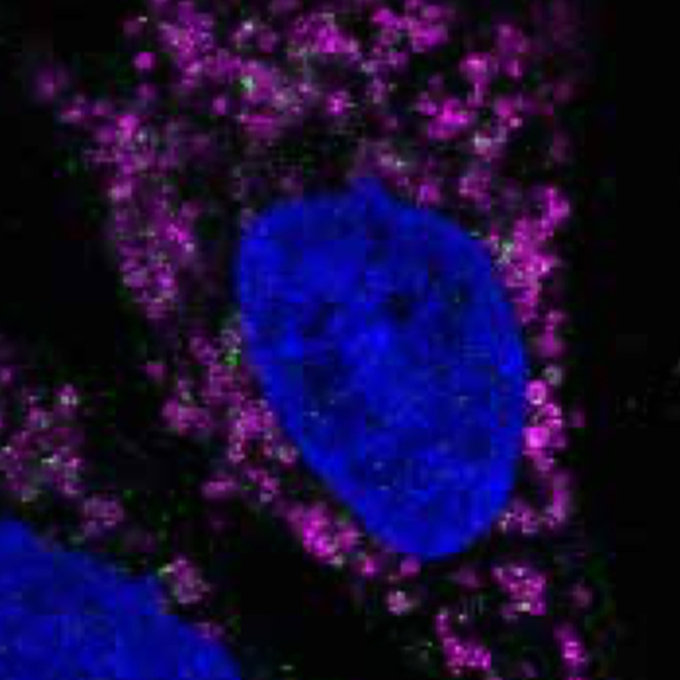
Nascent ribosomal RNA act as surfactant that suppresses growth of fibrillar centers in nucleolus
RNA Biofunction Laboratory
-
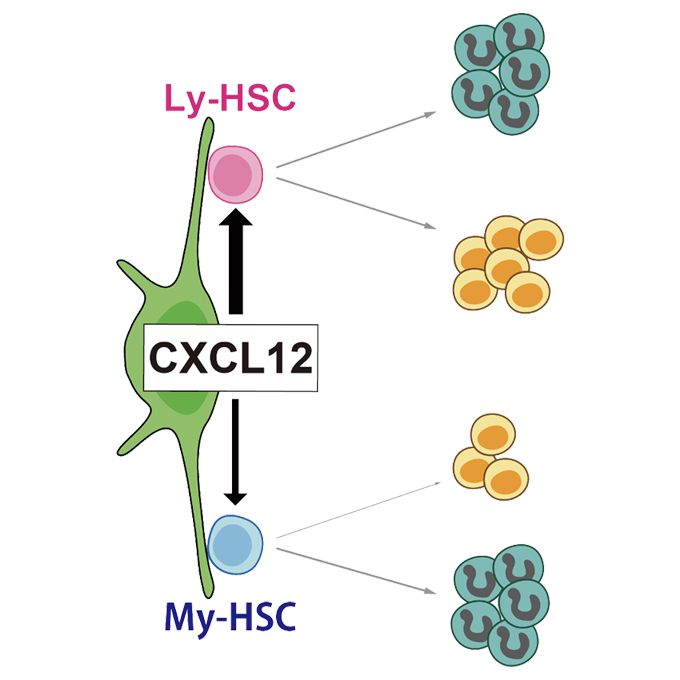
Ebf3+ niche-derived CXCL12 is required for the localization and maintenance of hematopoietic stem cells
Laboratory of Stem Cell Biology and Developmental Immunology
-
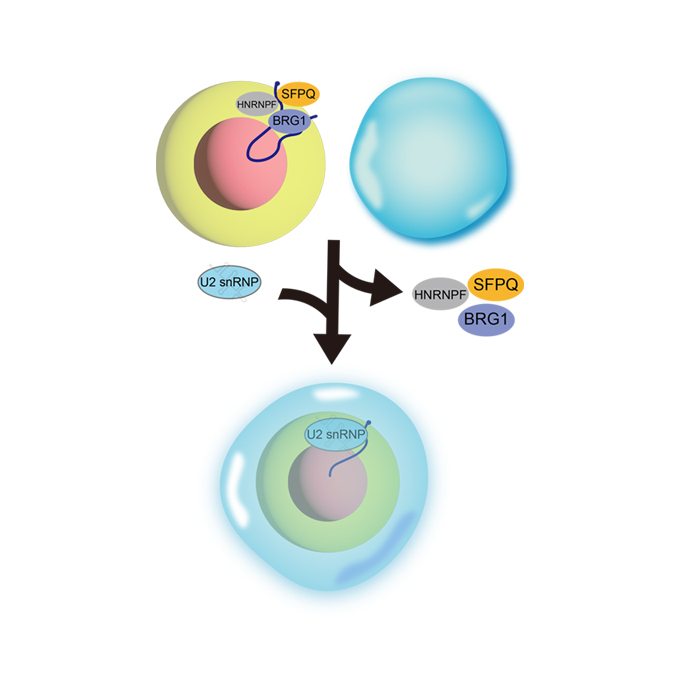
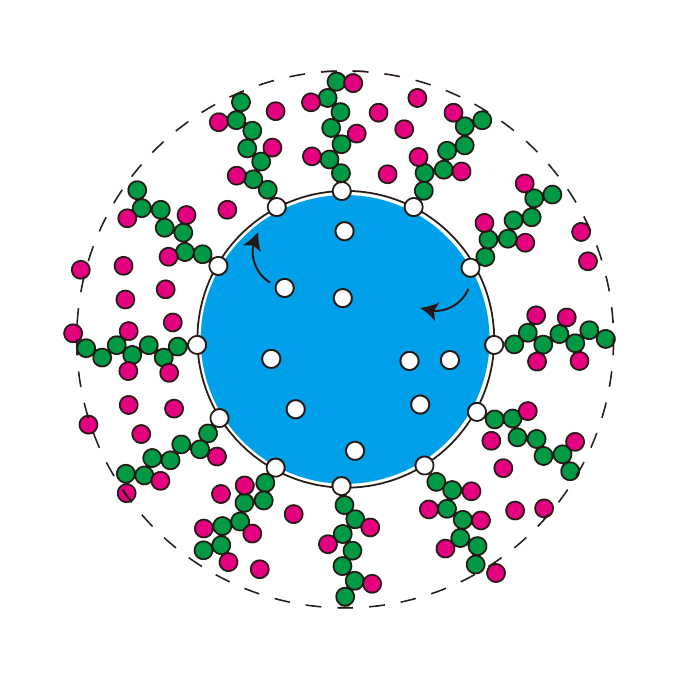
Shell protein composition specified by the lncRNA NEAT1 domains dictates the formation of paraspeckles as membraneless organelles
RNA Biofunction Laboratory
-
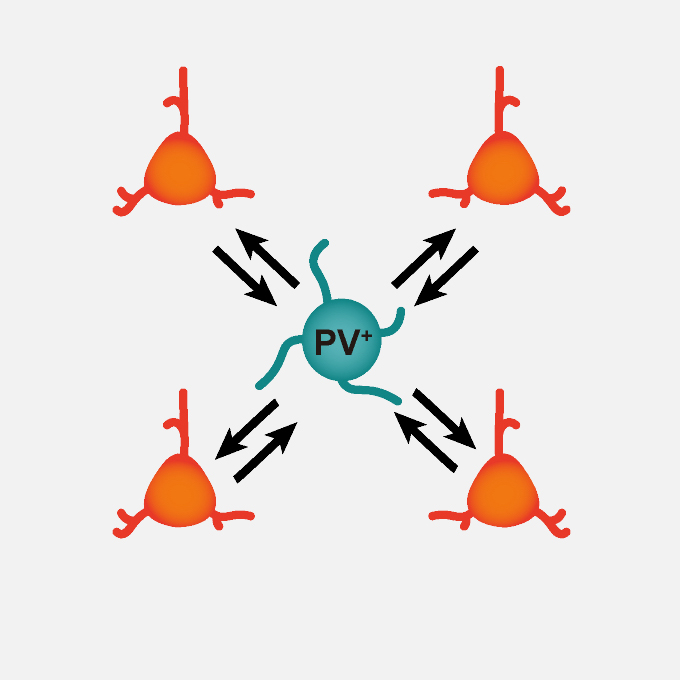
Reciprocal connections between parvalbumin-expressing 1 cells and adjacent 2 pyramidal cells are regulated by clustered protocadherin gamma
KOKORO-Biology Group
-
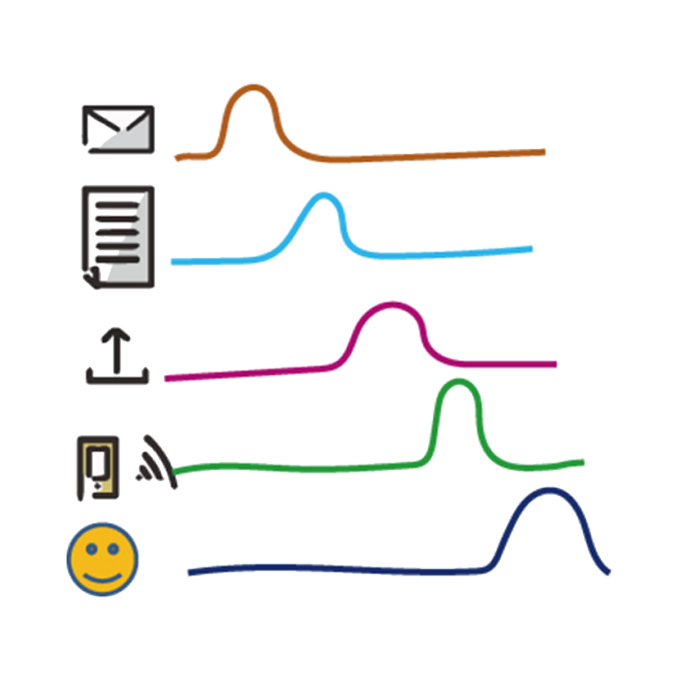
Cycles of goal silencing and reactivation underlie complex problem-solving in primate frontal and parietal cortex
Dynamic Brain Network Laboratory
-
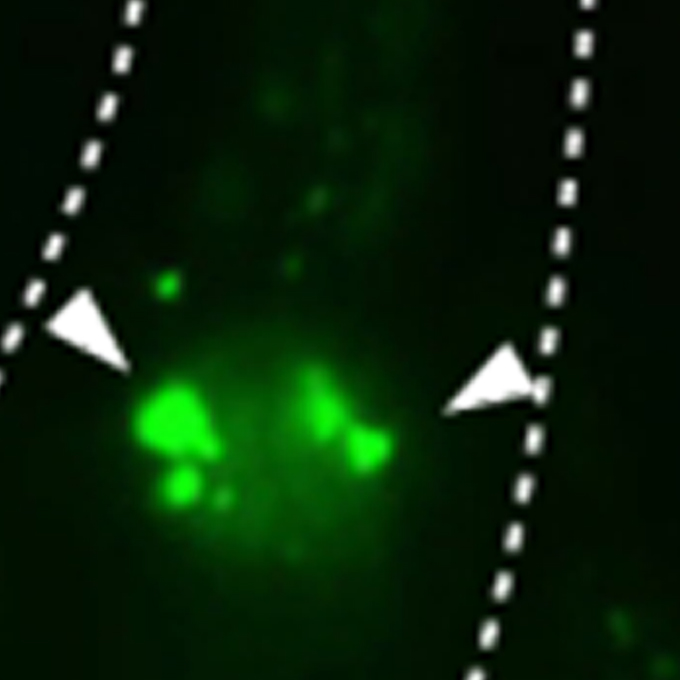
Neuronal MML-1/MXL-2 regulates systemic aging via glutamate transporter and cell non-autonomous autophagic and peroxidase activity
Laboratory of Intracellular Membrane Dynamics
-
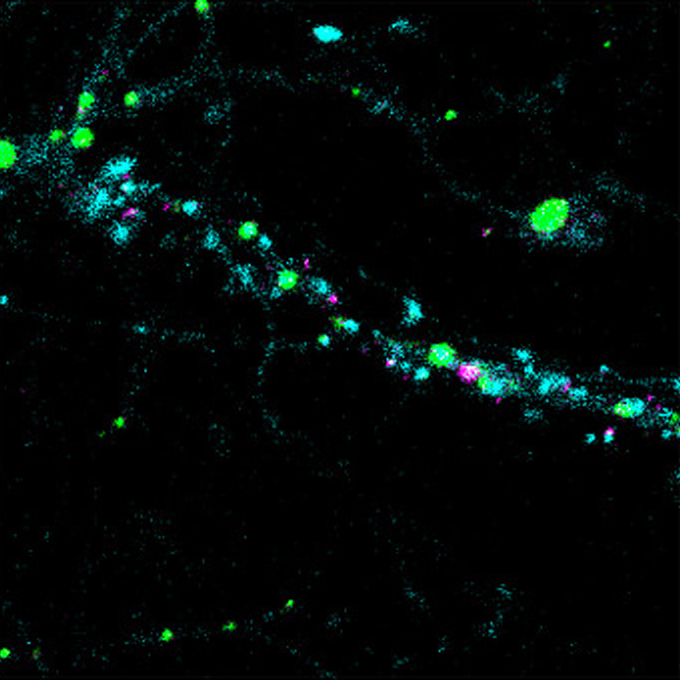
Visualization of trans homophilic interaction of clustered protocadherin in neurons
KOKORO-Biology Group
-
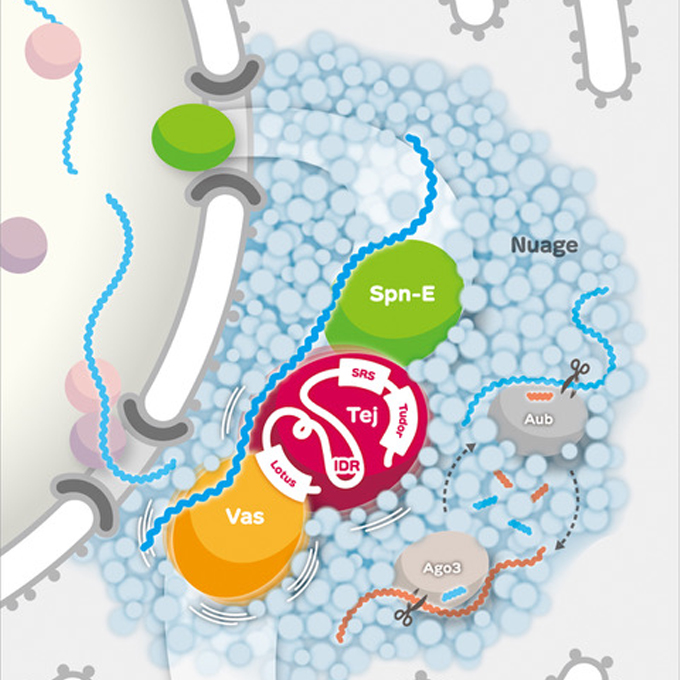
The protein protectors of fertility
Germline Biology Group
-
Satellite RNAs: emerging players in subnuclear architecture and gene regulation
RNA Biofunction Laboratory
-
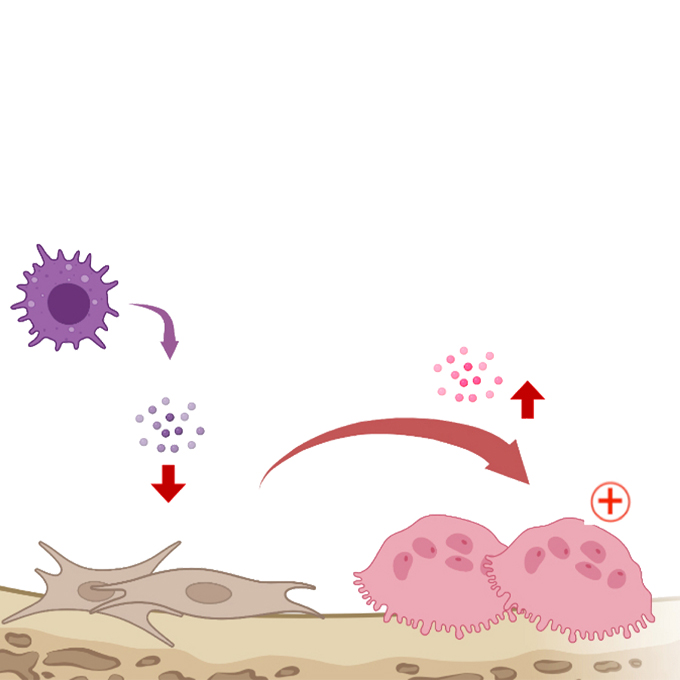
Ear today, gone tomorrow? A new discovery in a cause of inner-ear bone loss
Laboratory of Immunology and Cell Biology
-
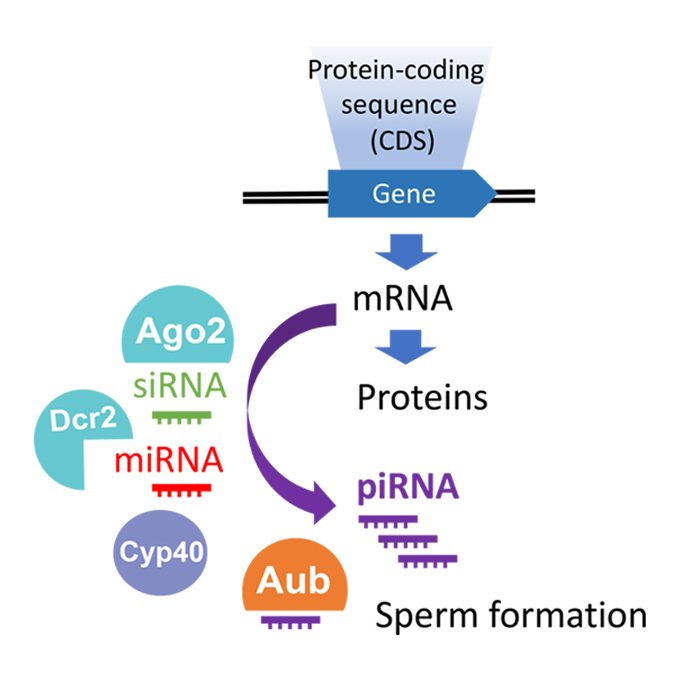
miRNA/siRNA-directed Pathway to Produce Non-coding piRNAs from Endogenous Protein-coding Regions Ensures Drosophila Spermatogenesis
Germline Biology Group
-
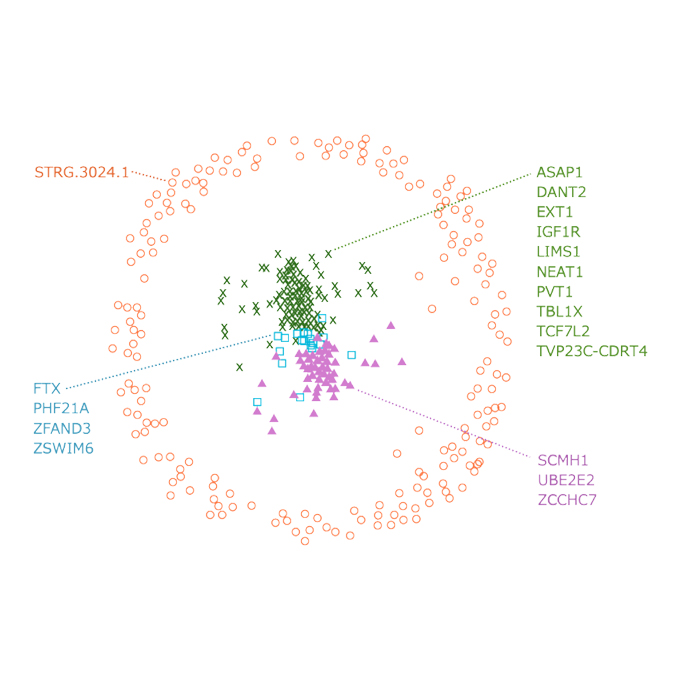
Unraveling the Mystery of Semi-extractable RNAs From Human Cell Lines
RNA Biofunction Laboratory
-
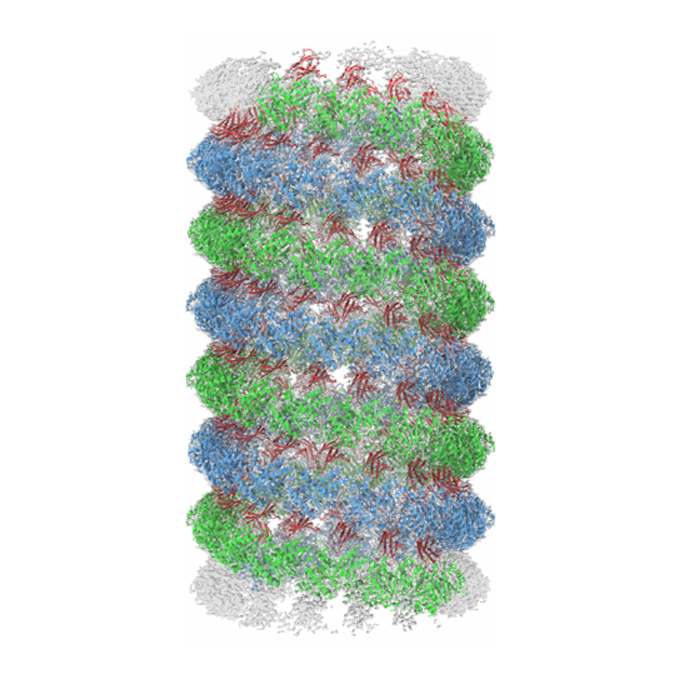
Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody
JEOL YOKOGUSHI Research Alliance Laboratories
-
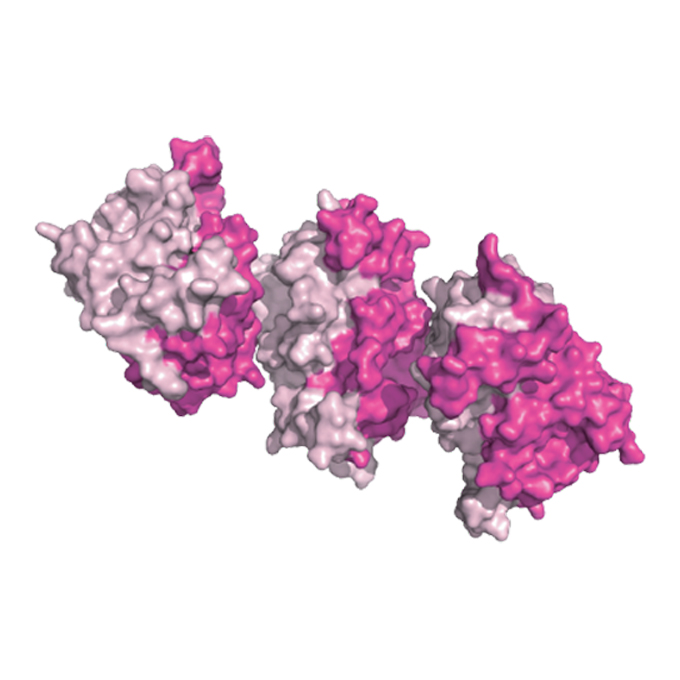
Pulling its weight: the protein key to chromosome movement during cell division
Laboratory of Chromosome Biology
-
CDYL reinforces male gonadal sex determination through epigenetically repressing Wnt4 transcription in mice
Laboratory of Epigenome Dynamics
-
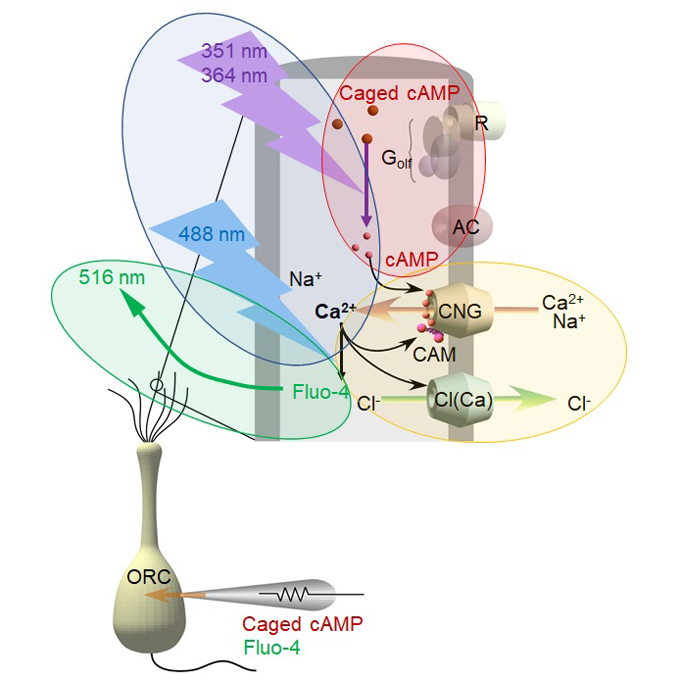
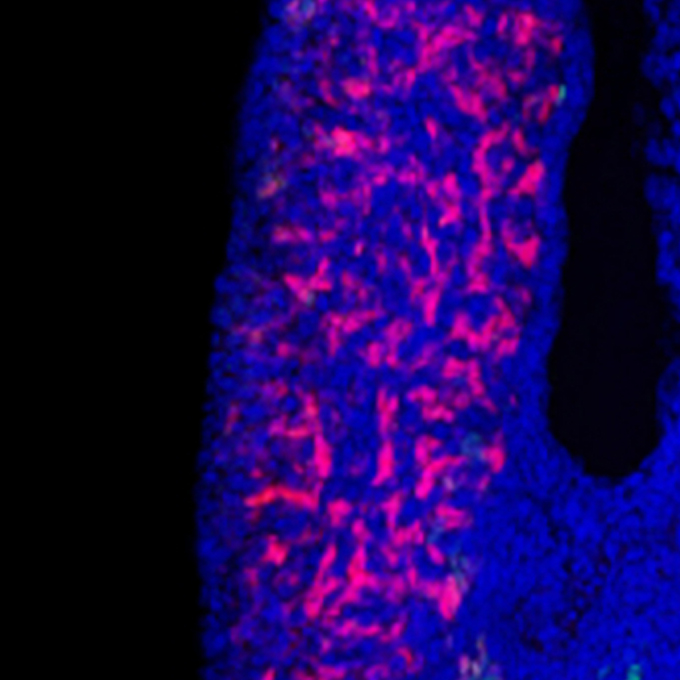
Can humans 'Sniff out' the secrets to the sense of smell?
Physiological Laboratory
-

Engraftment of allogeneic iPS cell-derived cartilage organoid in a primate model of articular cartilage defect
Laboratory of Tissue Biochemistry
-

Correlation between neural responses and human perception in figure-ground segregation
Cognitive Neuroscience Group
-
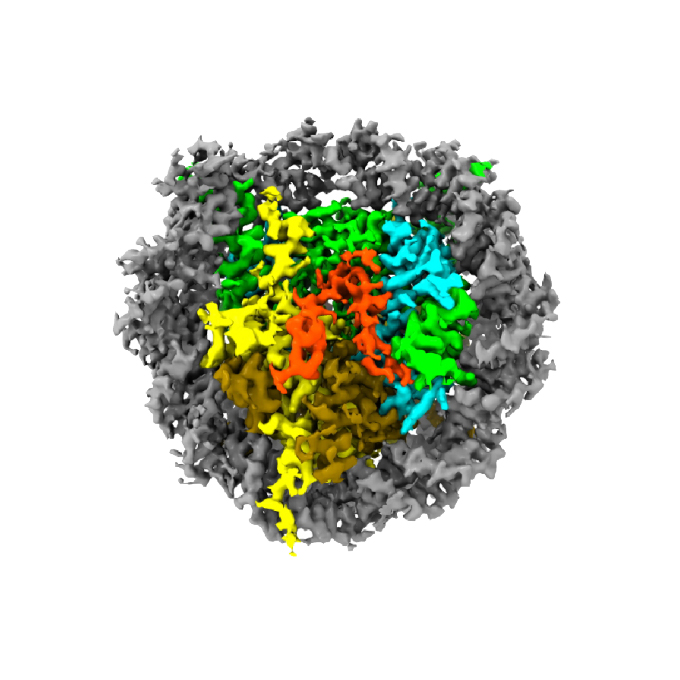
The cryo-EM structure of the CENP-A nucleosome in complex with ggKNL2
Laboratory of Chromosome Biology
-
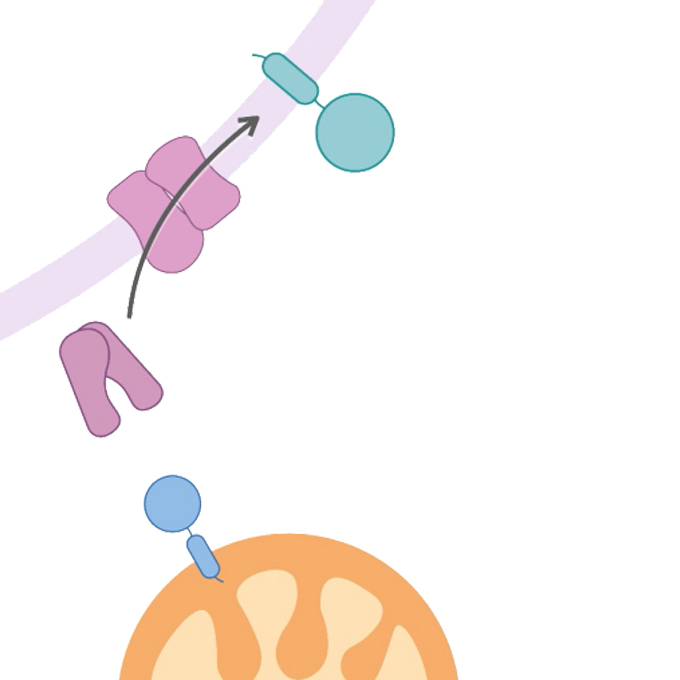
A cooperation between mitochondria and the endoplasmic reticulum is crucial for the mitochondrial health care system called mitophagy
Laboratory of Mitochondrial Dynamics
-
In vivo induction of activin A-producing alveolar macrophages supports the progression of lung cell carcinoma
Laboratory of Immunology and Cell Biology
2022
-
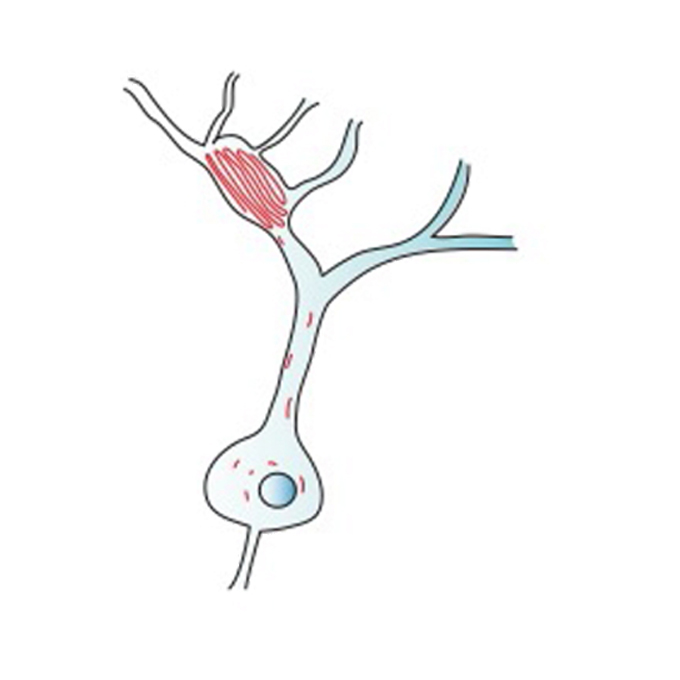
Isoform requirement of clustered protocadherin for preventing neuronal apoptosis and neonatal lethality
KOKORO-Biology Group
-
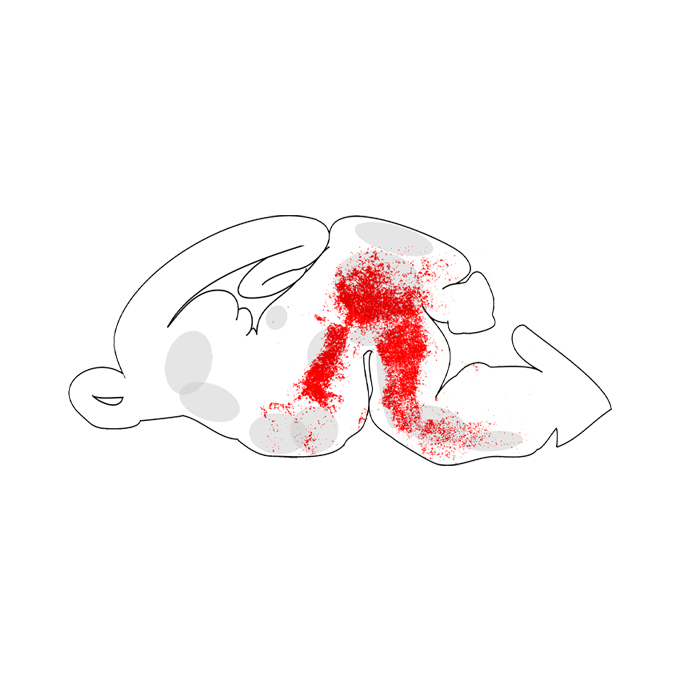
CTCF loss induces giant lamellar bodies in Purkinje cell dendrites
KOKORO-Biology Group
-
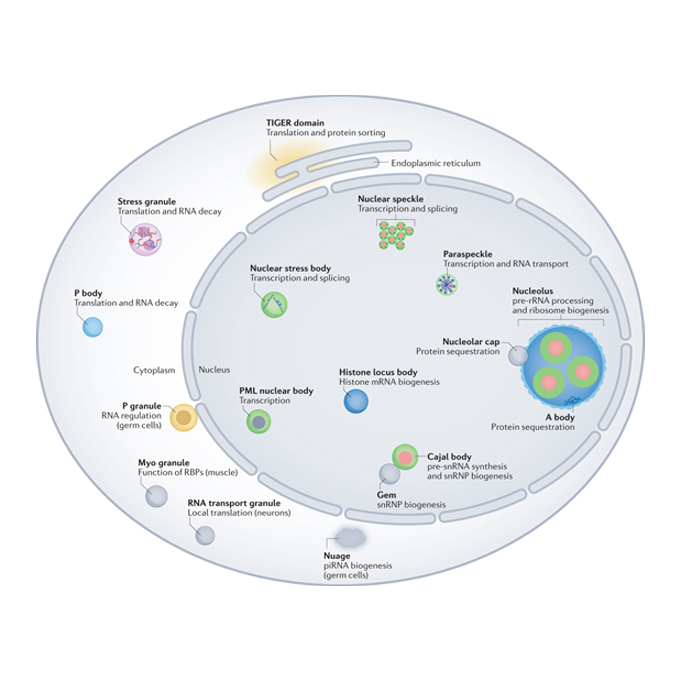
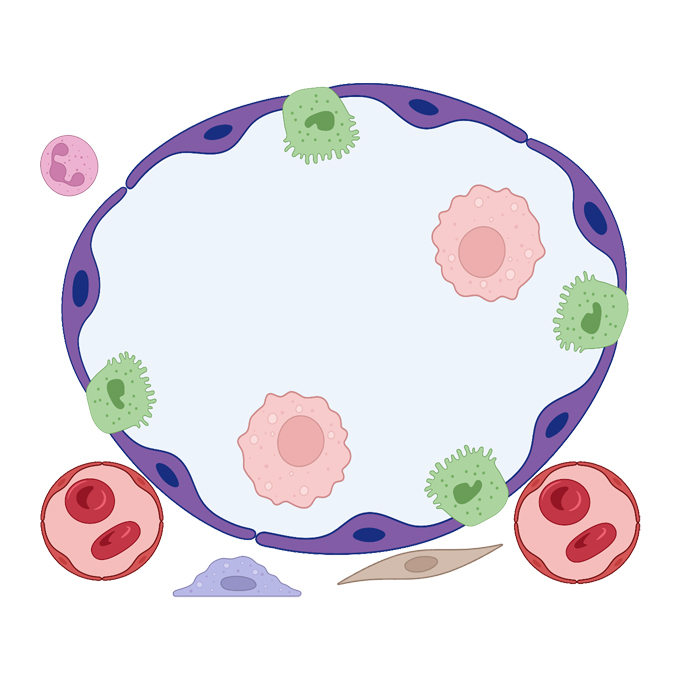
A guide to membraneless organelles and their various roles in gene regulation
RNA Biofunction Laboratory
-
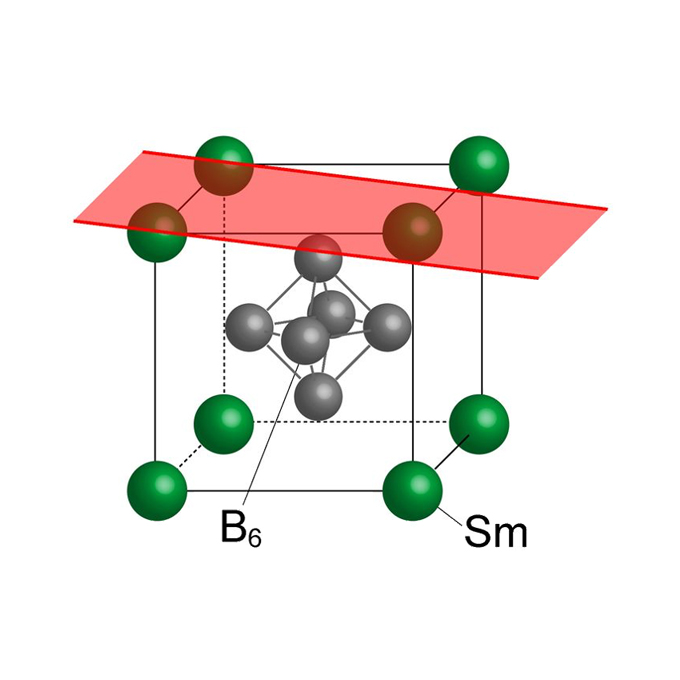
Breakdown of bulk-projected isotropy in surface electronic states of topological Kondo insulator SmB6(001)
Photophysics Laboratory
-
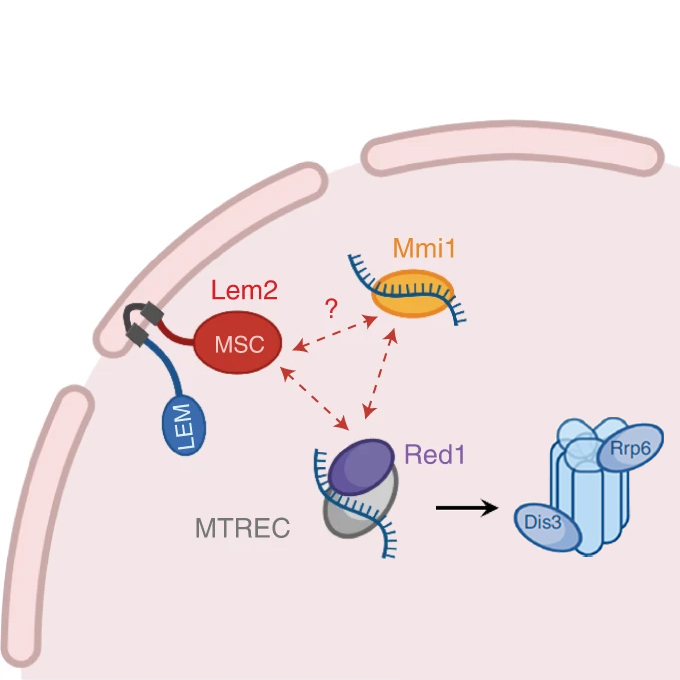
The inner nuclear membrane protein Lem2 coordinates RNA degradation at the nuclear periphery
Nuclear Dynamics Group
-
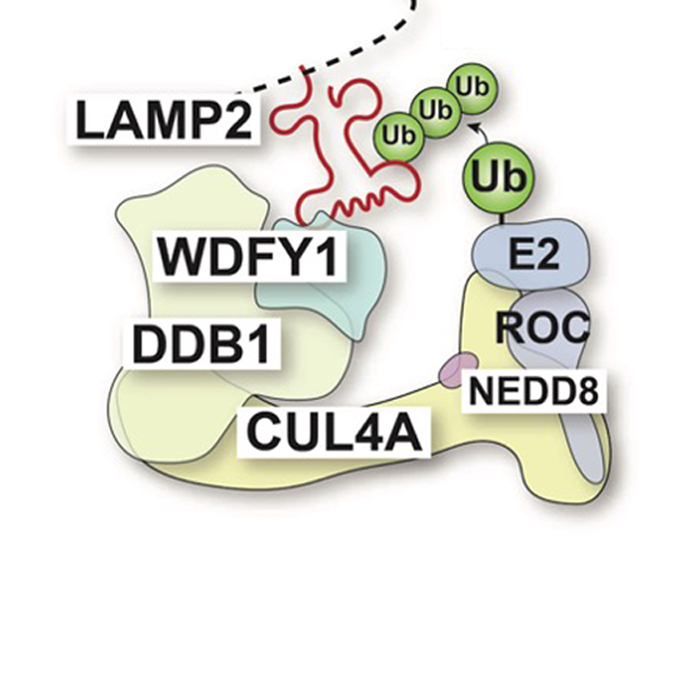
Identification of CUL4A- DDB1-WDFY1 as the E3 ubiquitin ligase complex to initiate lysophagy
Laboratory of Intracellular Membrane Dynamics
-
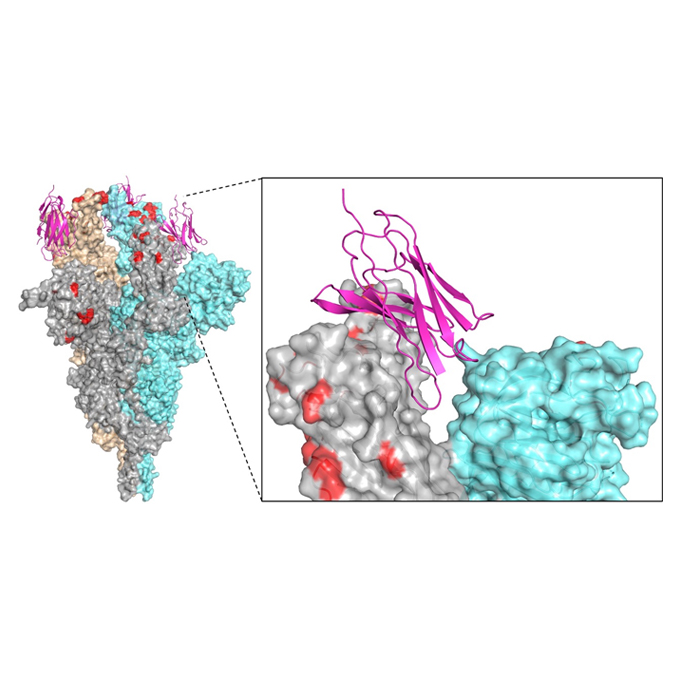
A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron
JEOL YOKOGUSHI Research Alliance Laboratories
-
You trust a face like yours
Dynamic Brain Network Laboratory
-
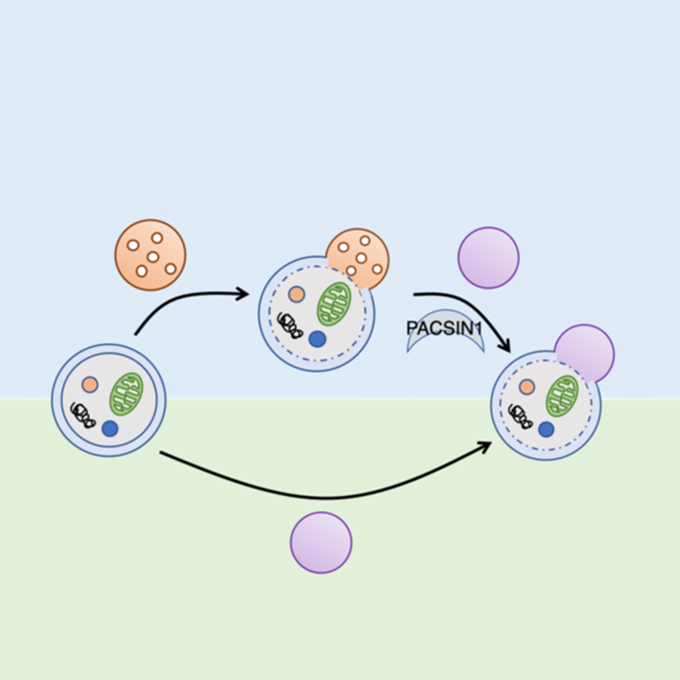
PACSIN1 is indispensable for amphisome-lysosome fusion during basal autophagy and subsets of selective autophagy
Laboratory of Intracellular Membrane Dynamics
-
Runx1 and Runx2 inhibit fibrotic conversion of cellular niches for hematopoietic stem cells
Laboratory of Stem Cell Biology and Developmental Immunology
-
Early embryonic mutations reveal dynamics of somatic and germ cell lineages in mice
KOKORO-Biology Group
-
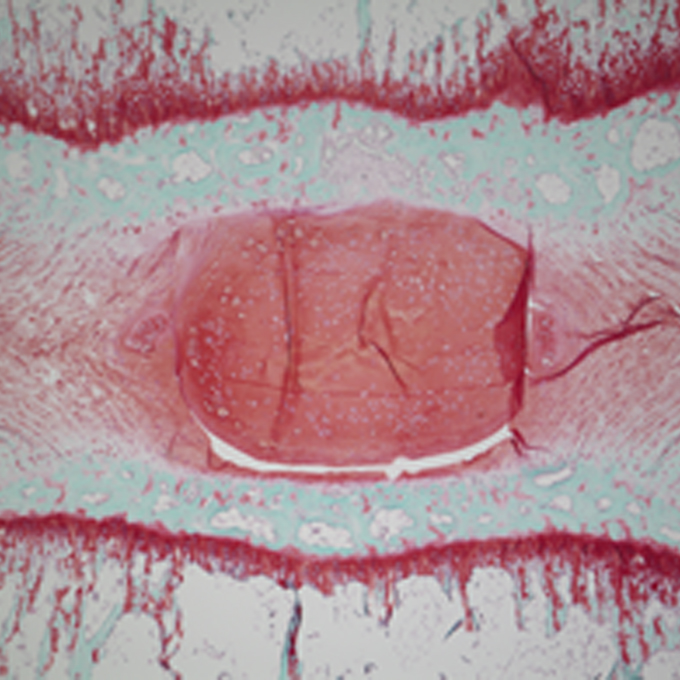
Human iPS cell-derived cartilaginous tissue spatially and functionally replaces nucleus pulposus
Laboratory of Tissue Biochemistry
-
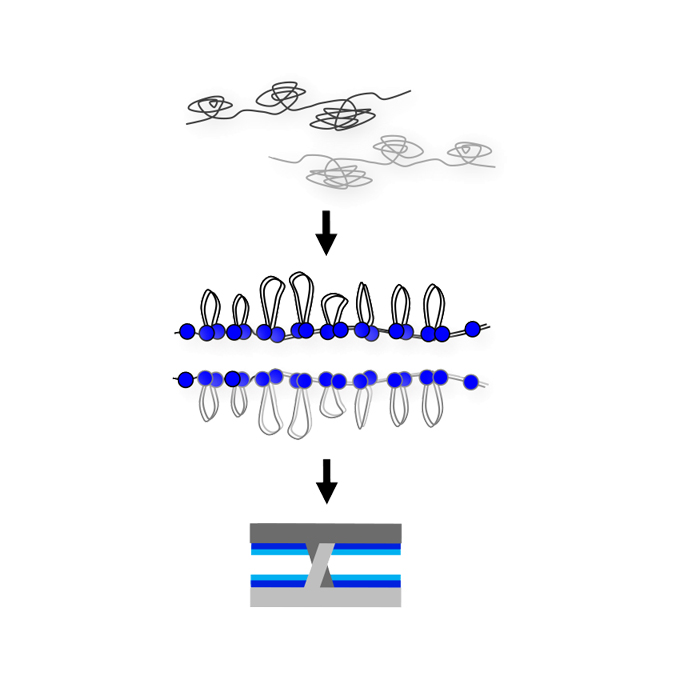
Rec8 cohesin-mediated axis-loop chromatin architecture is required for meiotic recombination
Nuclear Dynamics Group
-
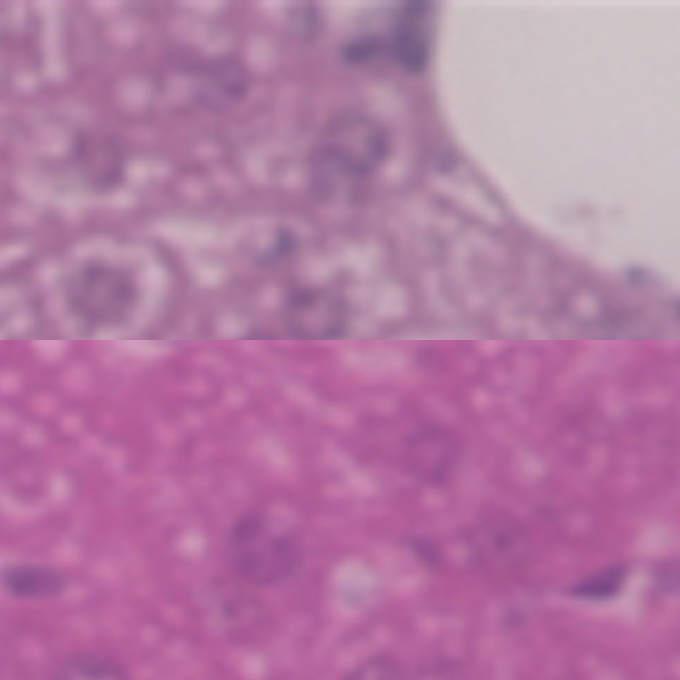
Loss of RUBCN/rubicon in adipocytes mediates the upregulation of autophagy to promote the fasting response
Laboratory of Intracellular Membrane Dynamics
-
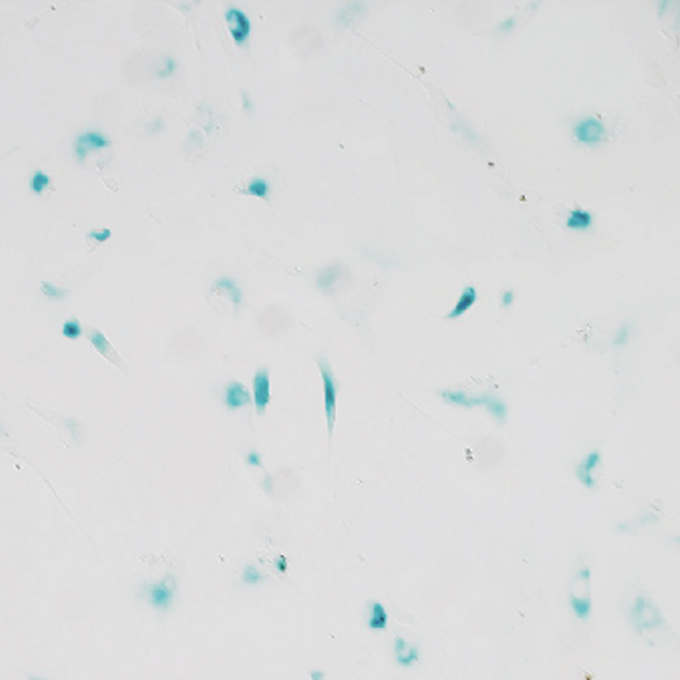
Age-associated decline of MondoA drives cellular senescence through impaired autophagy and mitochondrial homeostasis
Laboratory of Intracellular Membrane Dynamics
-
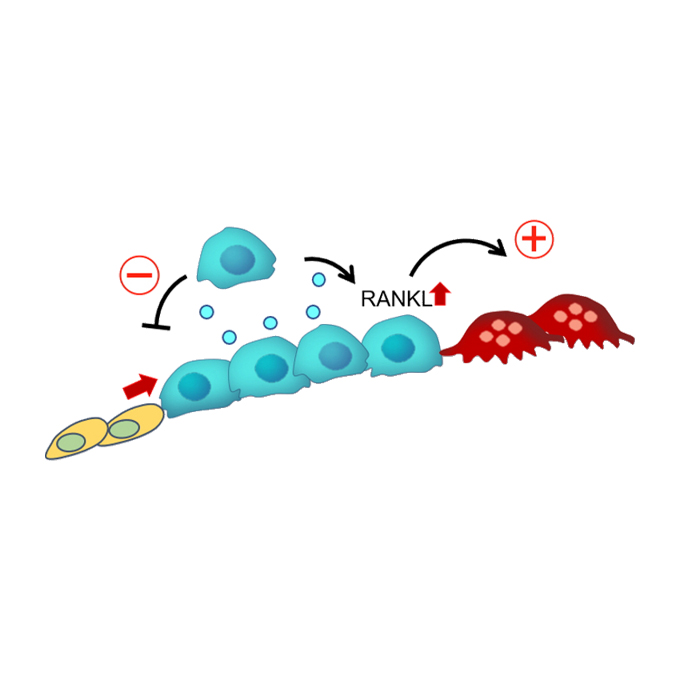
Osteoblast-derived vesicles induce a switch from bone-formation to bone-resorption in vivo
Laboratory of Immunology and Cell Biology
-
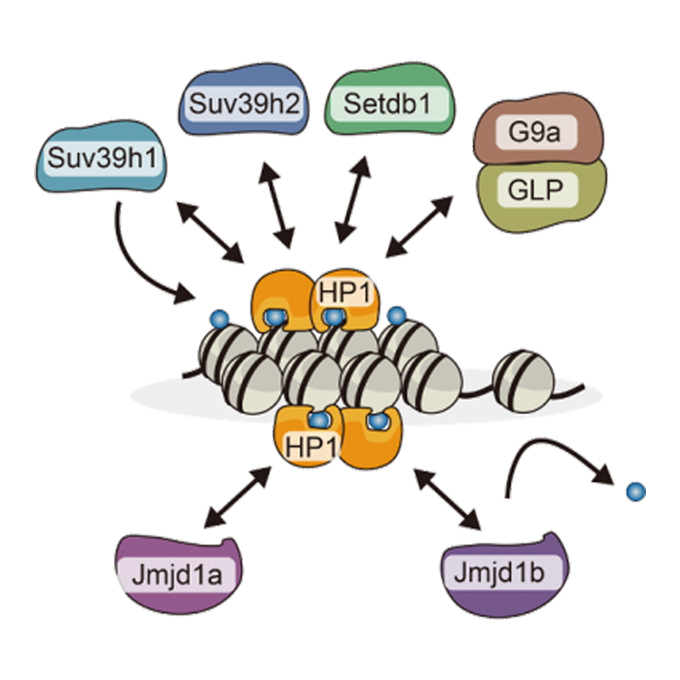
HP1 maintains protein stability of H3K9 methyltransferases and demethylases
Laboratory of Epigenome Dynamics
-
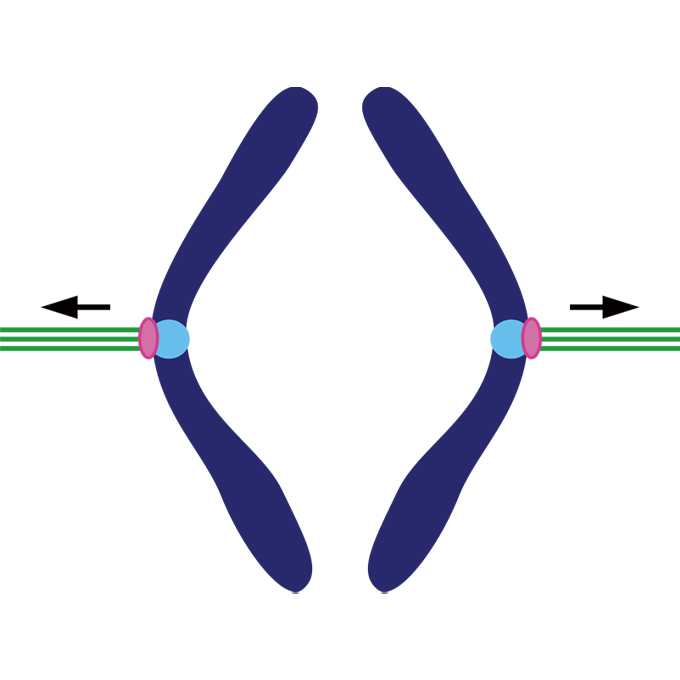
Recruitment of two Ndc80 complexes via the CENP-T pathway is sufficient for kinetochore functions
Laboratory of Chromosome Biology
-

Transfected plasmid DNA is incorporated into the nucleus via nuclear envelope reformation at telophase
Nuclear Dynamics Group
-
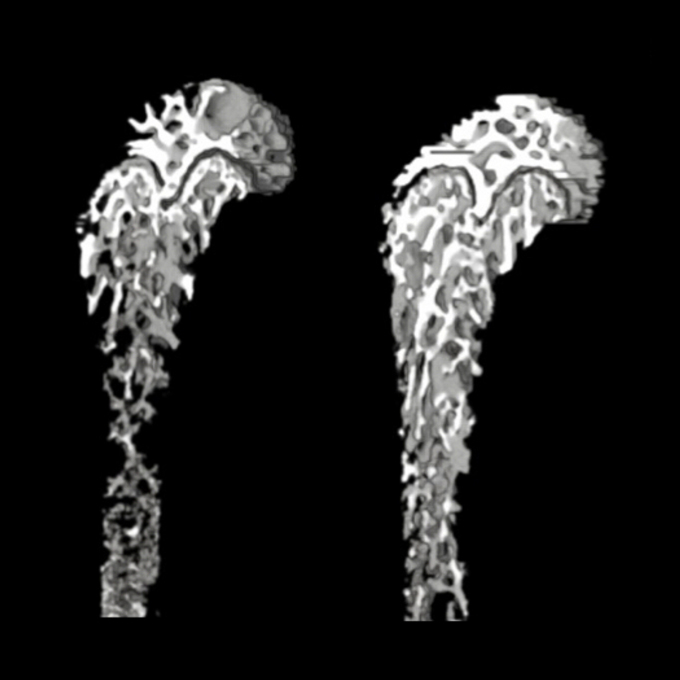
Degradation of the NOTCH intracellular domain by elevated autophagy in osteoblasts promotes osteoblast differentiation and alleviates osteoporosis
Laboratory of Intracellular Membrane Dynamics
-
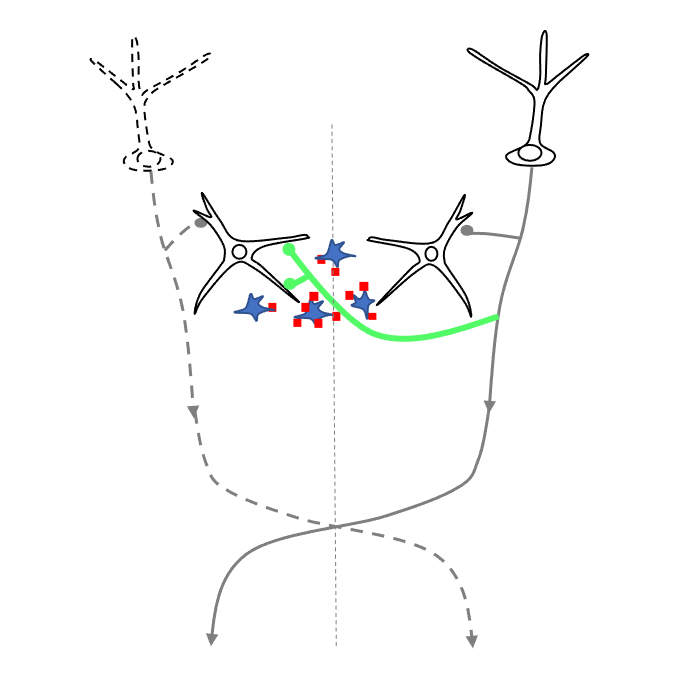
Involvement of denervated midbrain-derived factors in the formation of ectopic cortico-mesencephalic projection after hemispherectomy
Cellular and Molecular Neurobiology Group
2021
-
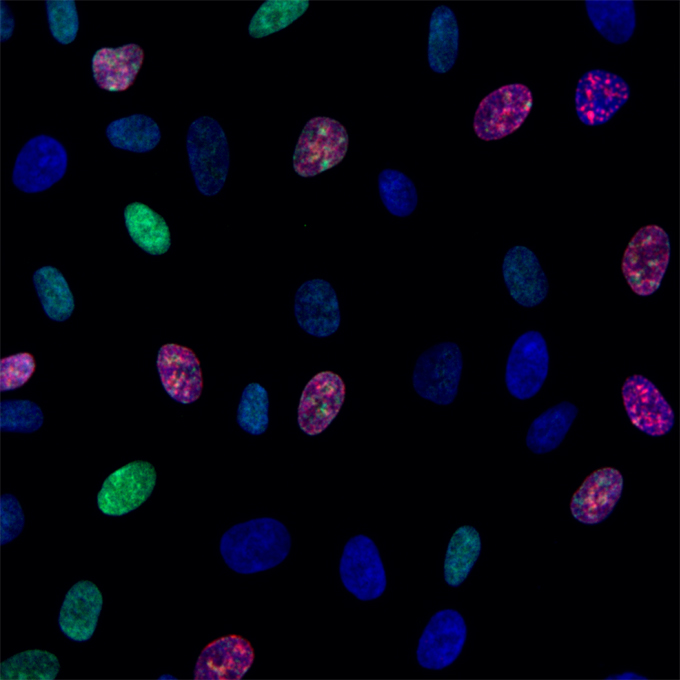
Chromatin loading of MCM hexamers is associated with di-/tri-methylation of histone H4K20 toward S phase entry
Nuclear Dynamics Group
-
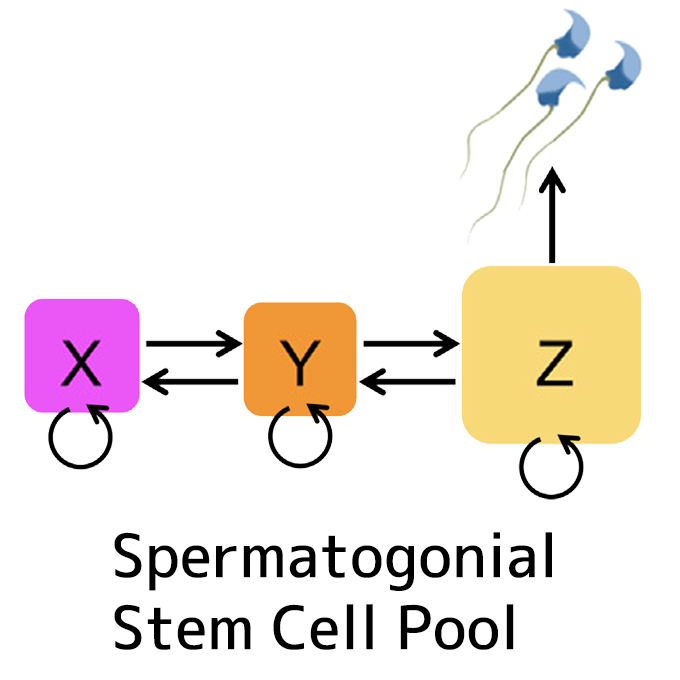
A multistate stem cell dynamics maintains homeostasis in mouse spermatogenesis
Laboratory of Stem Cell Biology and Developmental Immunology
-
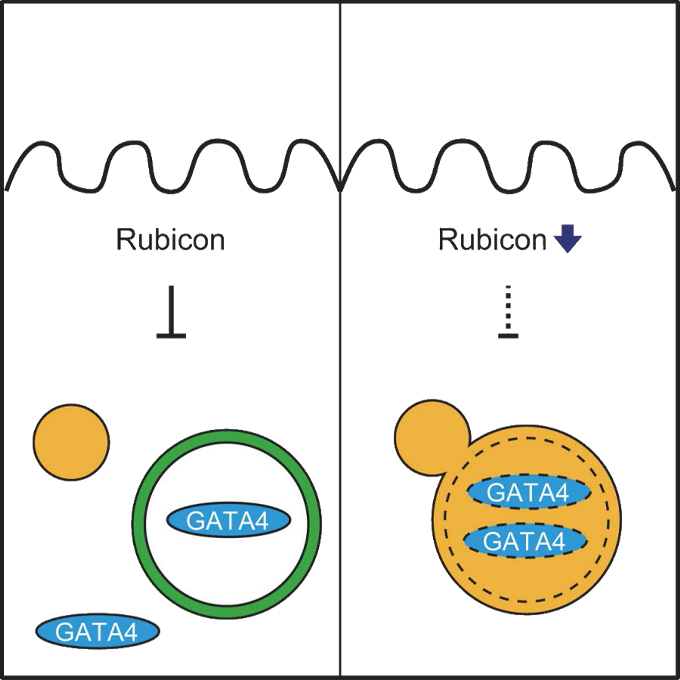
Rubicon prevents autophagic degradation of GATA4 to promote Sertoli cell function
Laboratory of Intracellular Membrane Dynamics
-
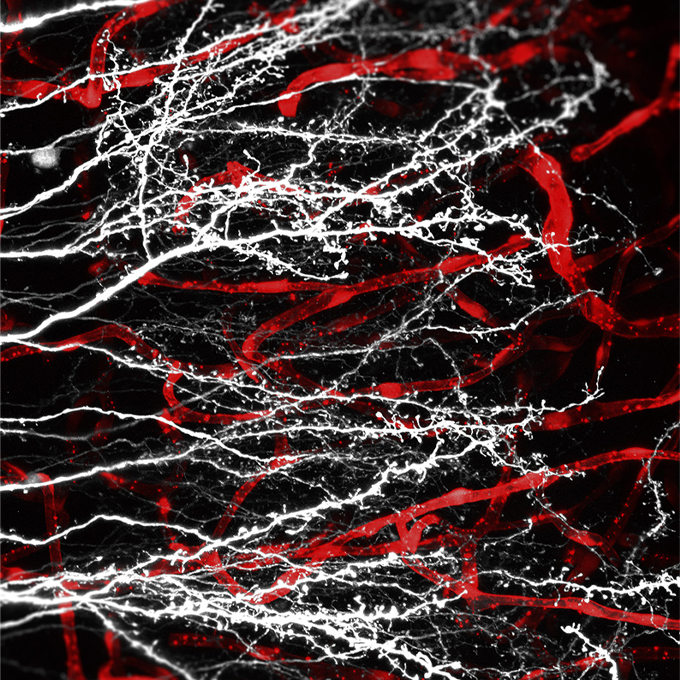
Ras-like Gem GTPase induced by Npas4 promotes activity-dependent neuronal tolerance for ischemic stroke
Cellular and Molecular Neurobiology Group
-
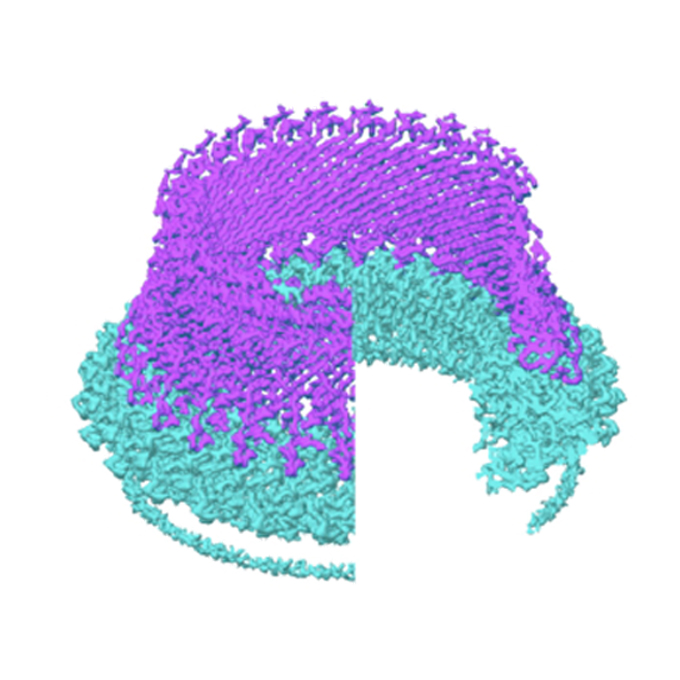
Structure of the molecular bushing of the bacterial flagellar motor
JEOL YOKOGUSHI Research Alliance Laboratories
-
Pivot burrowing of scarab beetle (Trypoxylus dichotomus) larva
Laboratory of Pattern Formation
-

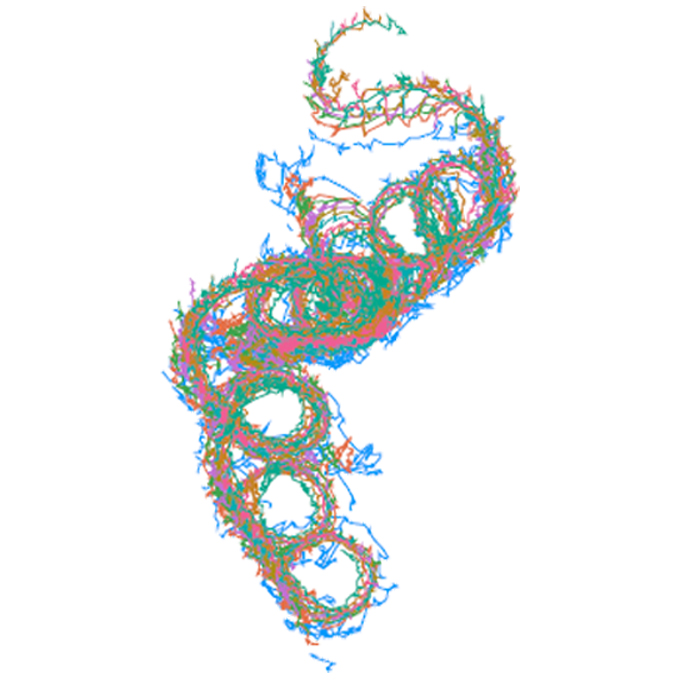
Native flagellar MS ring is formed by 34 subunits with 23-fold and 11-fold subsymmetries
JEOL YOKOGUSHI Research Alliance Laboratories
-
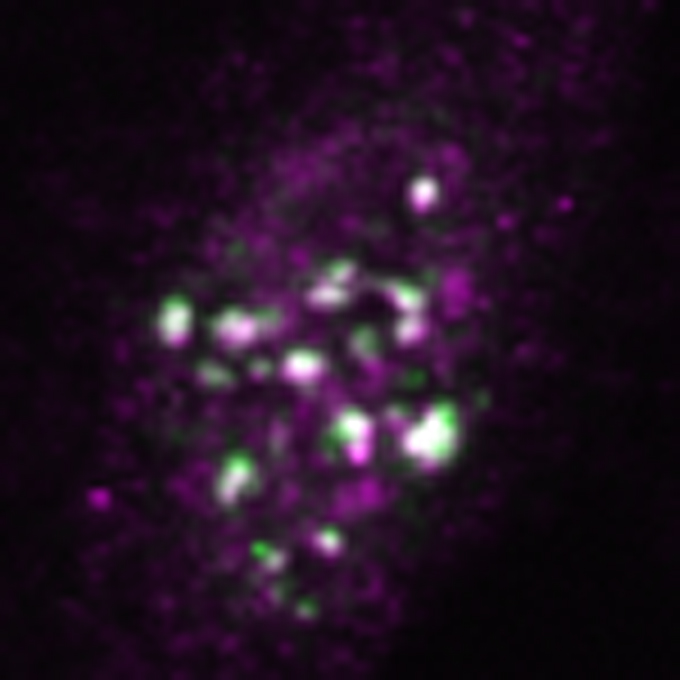
m6A-modification of HSATIII lncRNAs regulates temperature-dependent splicing
RNA Biofunction Laboratory
-
Planar cell polarity induces local microtubule bundling for coordinated ciliary beating
Laboratory of Barriology and Cell Biology
-
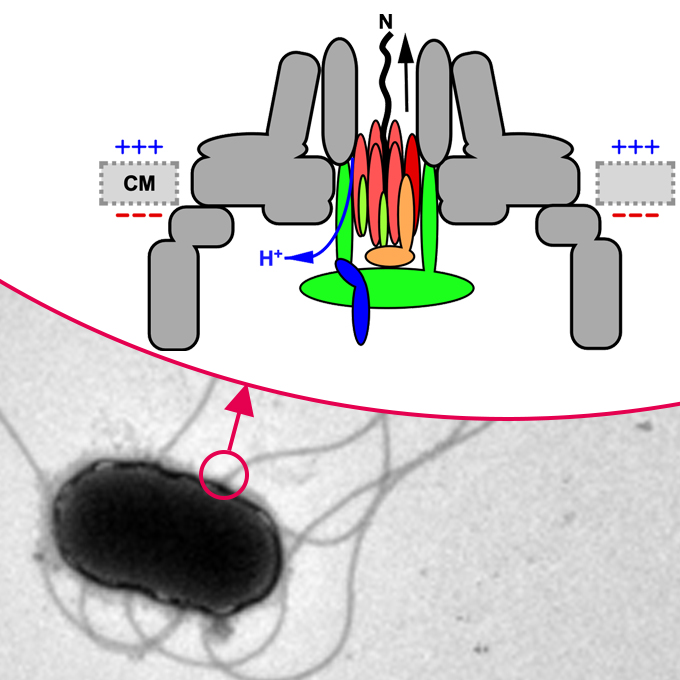
Membrane voltage-dependent activation mechanism of the bacterial flagellar protein export apparatus
JEOL YOKOGUSHI Research Alliance Laboratories
-
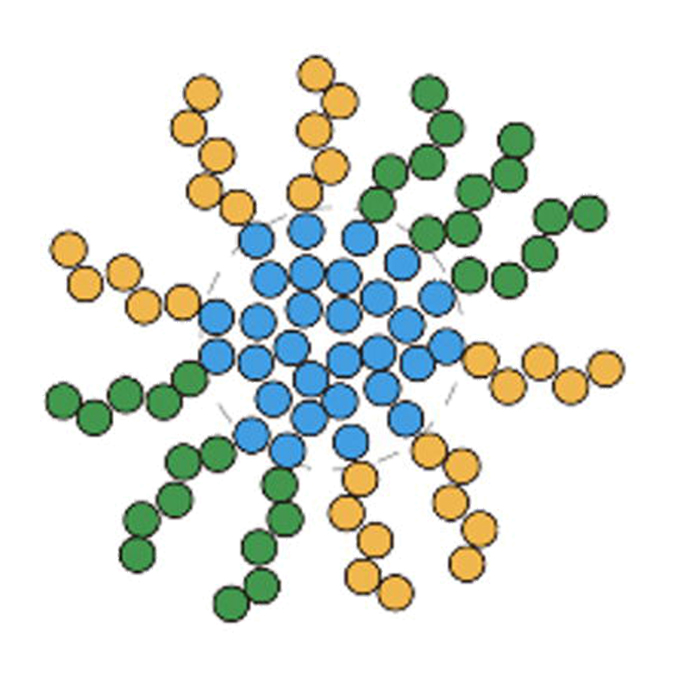
Paraspeckles are constructed as block copolymer micelles
RNA Biofunction Laboratory
-
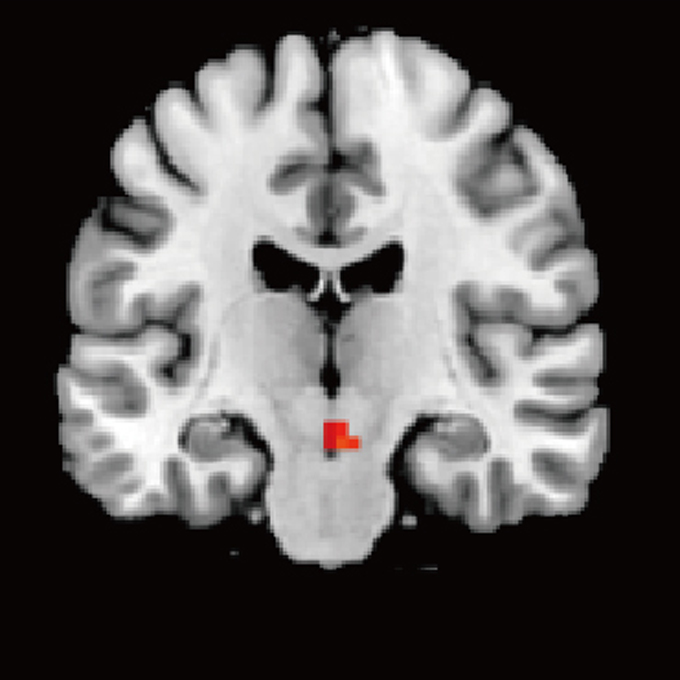
Self-face activates the dopamine reward pathway without awareness
Dynamic Brain Network Laboratory
-
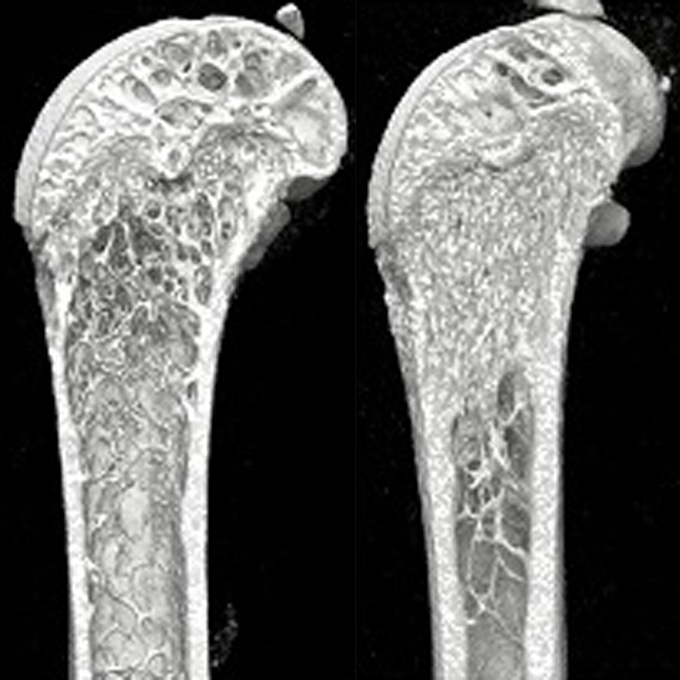
SLPI is a critical mediator that controls PTH-induced bone formation
Laboratory of Immunology and Cell Biology
-
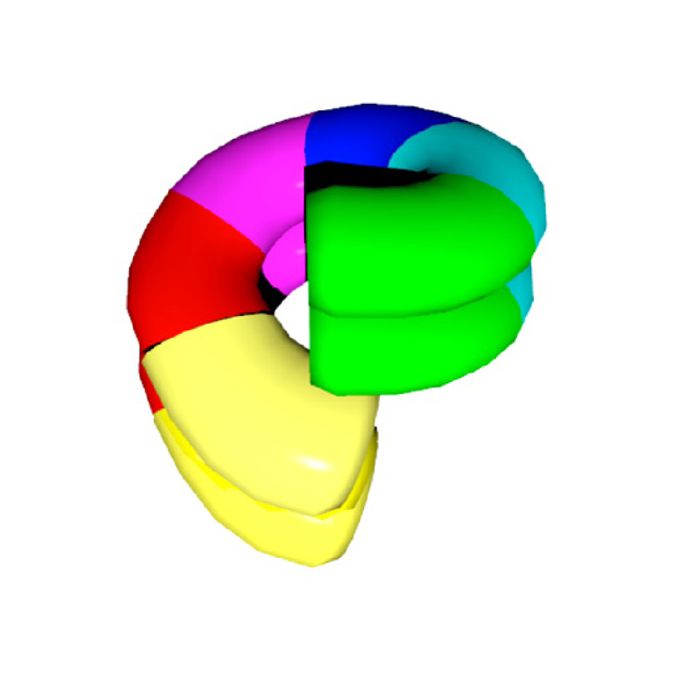
Split conformation of Chaetomium thermophilum Hsp104 disaggregase
JEOL YOKOGUSHI Research Alliance Laboratories
-
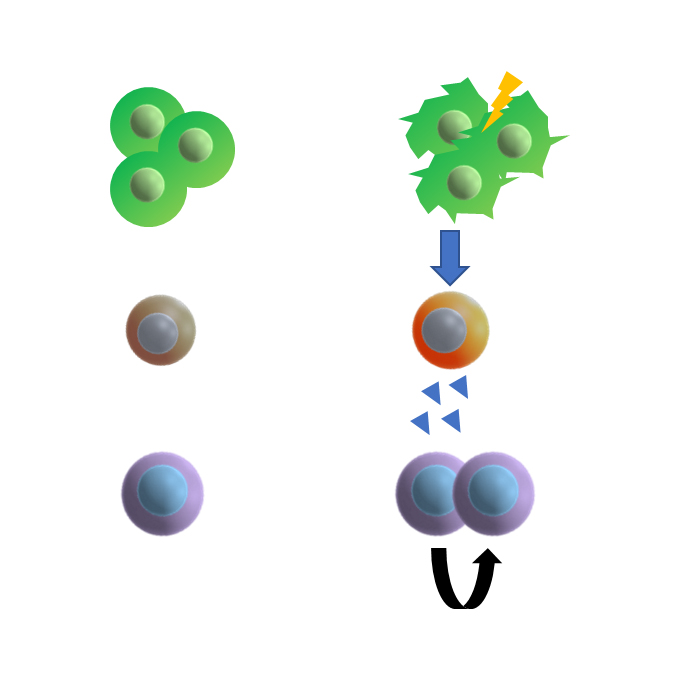
Group 2 innate lymphoid cells support hematopoietic recovery under stress conditions
Laboratory of Immunology and Cell Biology
-
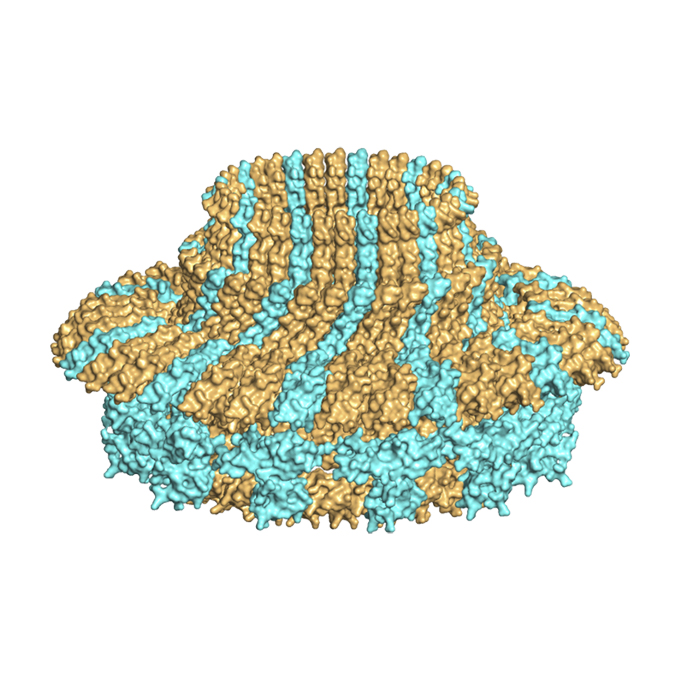
Two distinct conformations in 34 FliF subunits generate three different symmetries within the flagellar MS-ring
JEOL YOKOGUSHI Research Alliance Laboratories
-
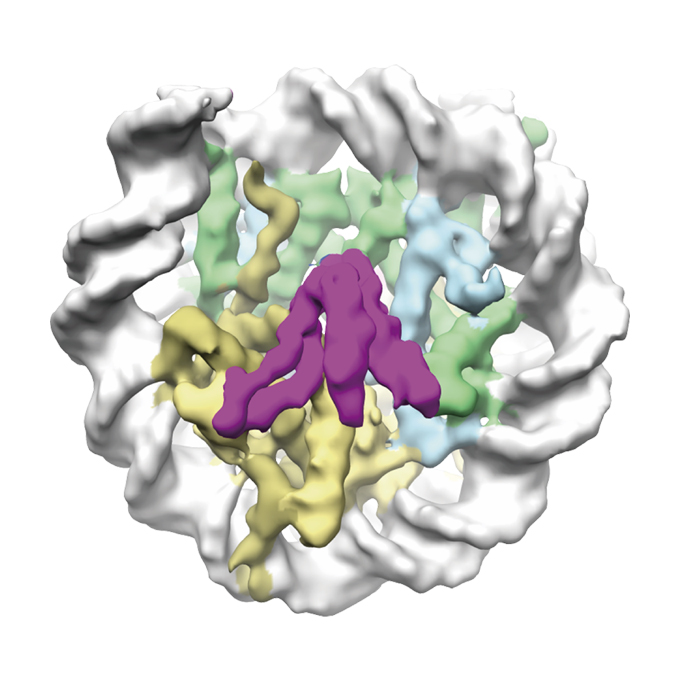
Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C
Laboratory of Chromosome Biology
-
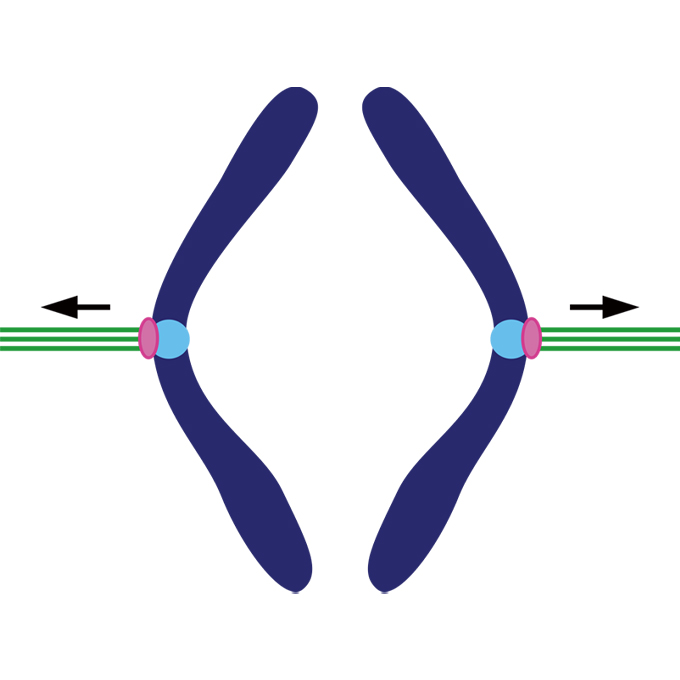
Bridgin connects the outer kinetochore to centromeric chromatin
Laboratory of Chromosome Biology
-
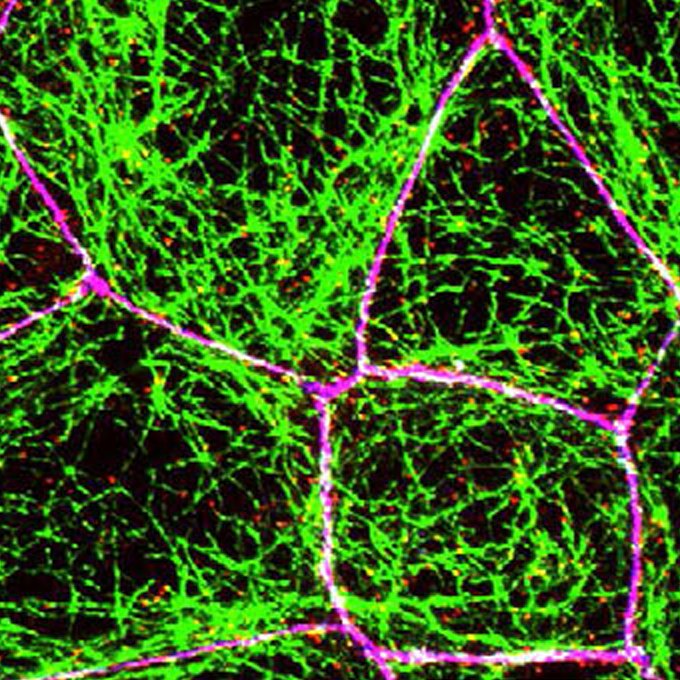
A microtubule-LUZP1 association around tight junction promotes epithelial cell apical constriction
Laboratory of Barriology and Cell Biology
2020
-
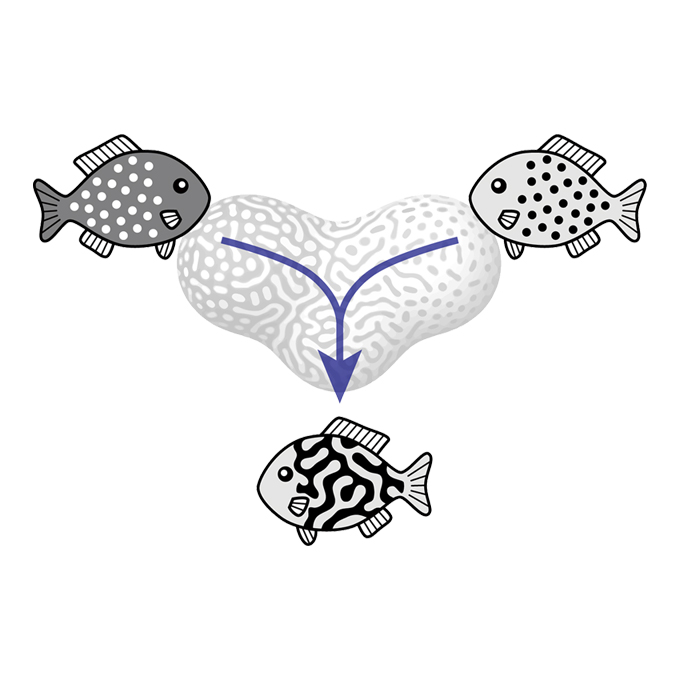
Pattern blending enriches the diversity of animal colorations
Laboratory of Pattern Formation
-
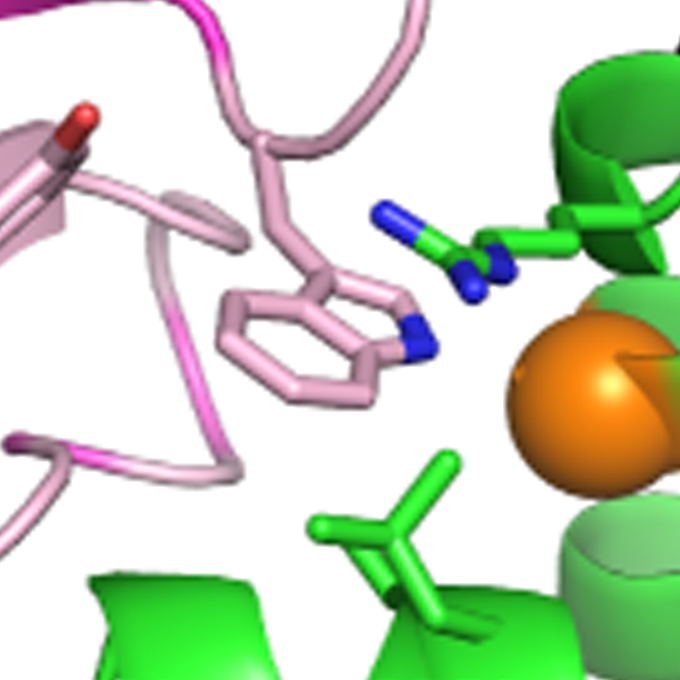
Essentiality of CENP-A depends on its binding mode to HJURP
Laboratory of Chromosome Biology
-
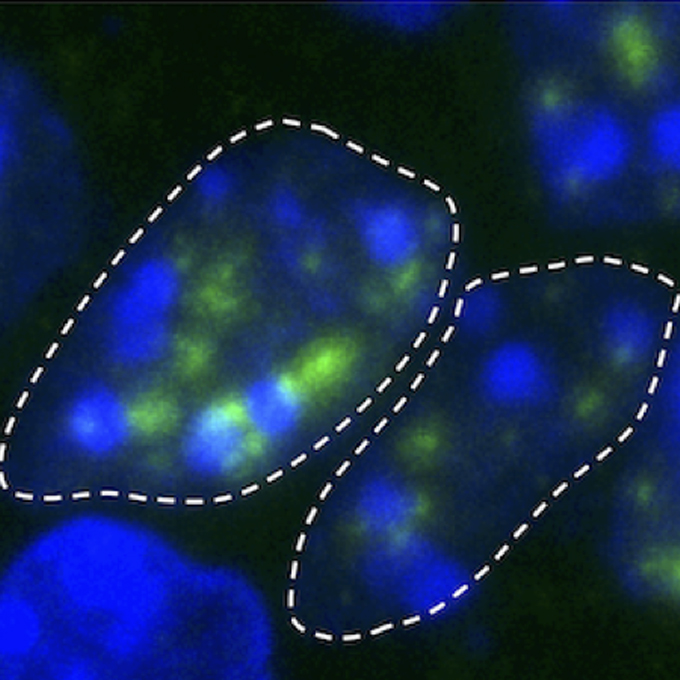
Modulation of Ago2 Loading by Cyclophilin 40 Endows a Unique Repertoire of Functional miRNAs during Sperm Maturation in Drosophila
Germline Biology Group
-
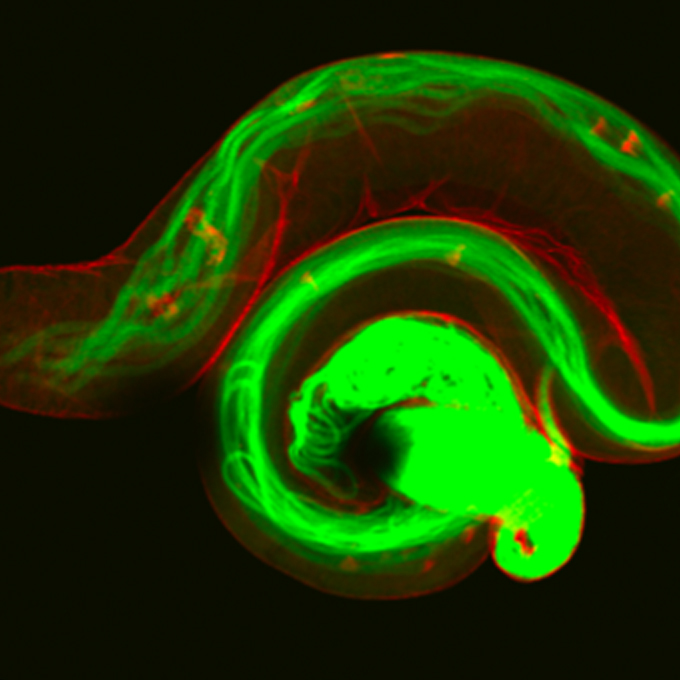
Suppression of DNA Double-Strand Break Formation by DNA Polymerase β in Active DNA Demethylation is Required for Development of Hippocampal Pyramidal Neurons
-
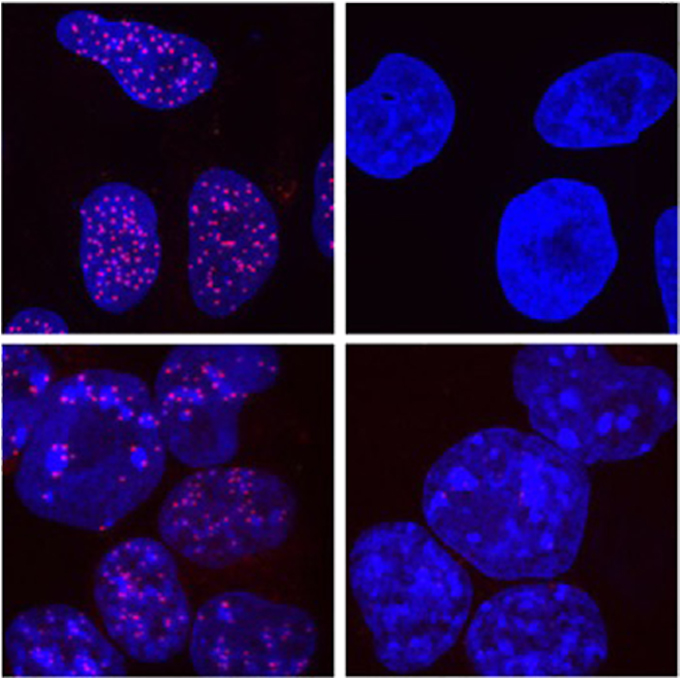
A super-sensitive auxin-inducible degron system with an engineered auxin-TIR1 pair
Laboratory of Chromosome Biology
-
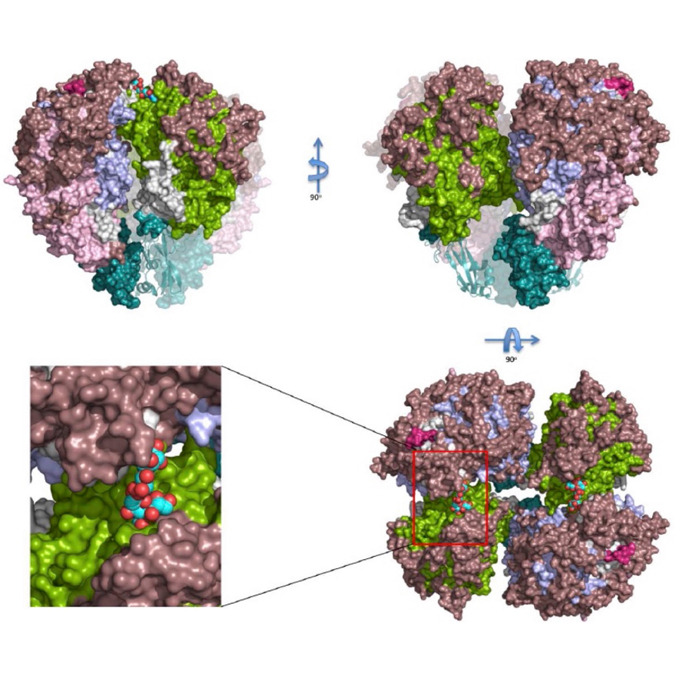
Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae
JEOL YOKOGUSHI Research Alliance Laboratories
-
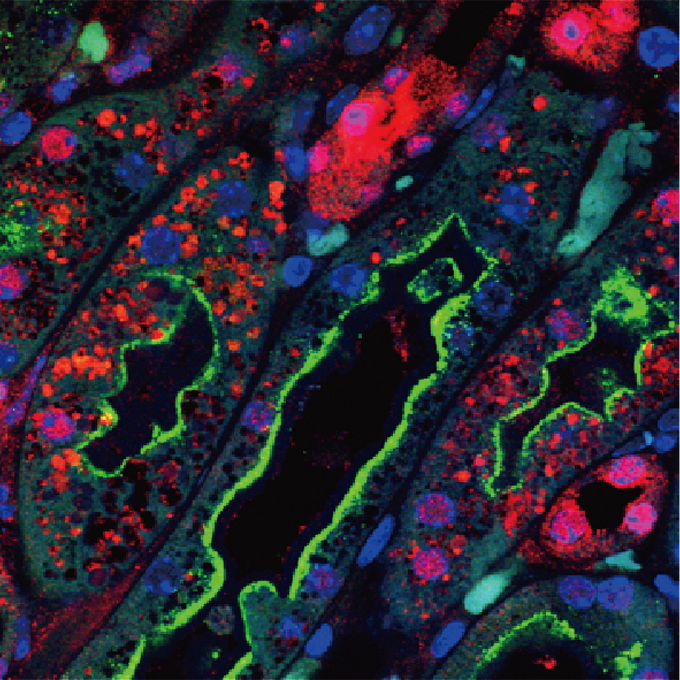
LC3 lipidation is essential for TFEB activation during the lysosomal damage response to kidney injury
Laboratory of Intracellular Membrane Dynamics
-

The mouse Sry locus harbors a cryptic exon that is essential for male sex determination
Laboratory of Epigenome Dynamics
-

Age-dependent loss of adipose Rubicon promotes metabolic disorders via excess autophagy
Laboratory of Intracellular Membrane Dynamics
-
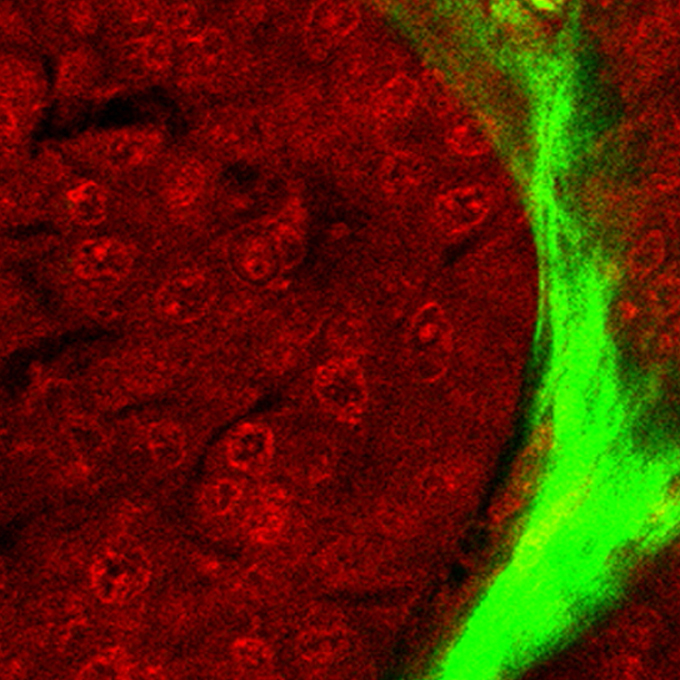
Nonlinear optics with near-infrared excitation enable real-time quantitative diagnosis of human cervical cancers
Laboratory of Immunology and Cell Biology
-
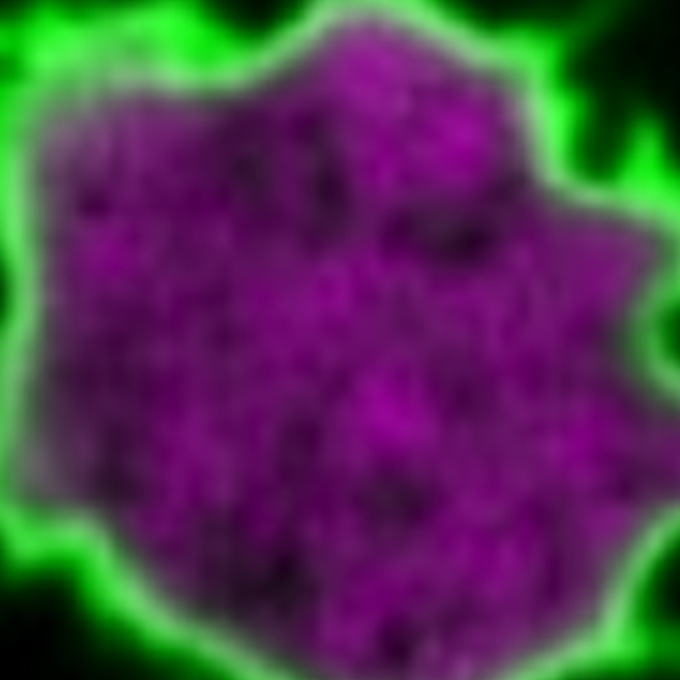
Single-molecule imaging of PI(4,5)P2 and PTEN in vitro reveals a positive feedback mechanism for PTEN membrane binding
Laboratory of Single Molecule Biology
-
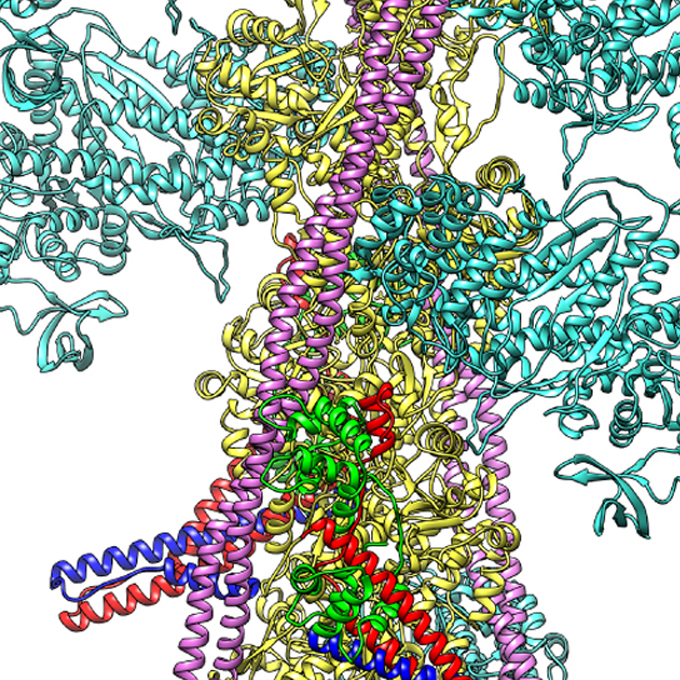
Getting to the "Heart" of Heart Beats: Cardiac thin filament structure and function revealed
JEOL YOKOGUSHI Research Alliance Laboratories
2019
-
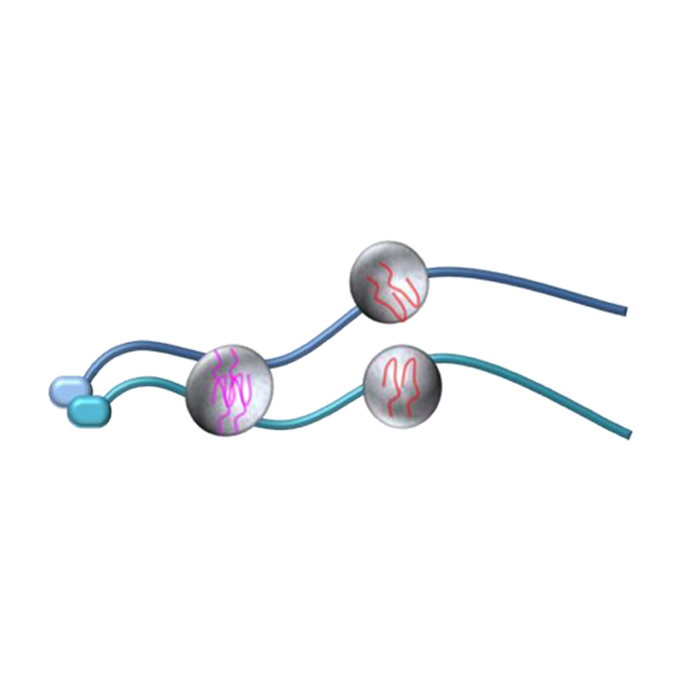
Chromosome-associated RNA-protein complexes promote pairing of homologous chromosomes during meiosis in Schizosaccharomyces pombe
Nuclear Dynamics Group
-
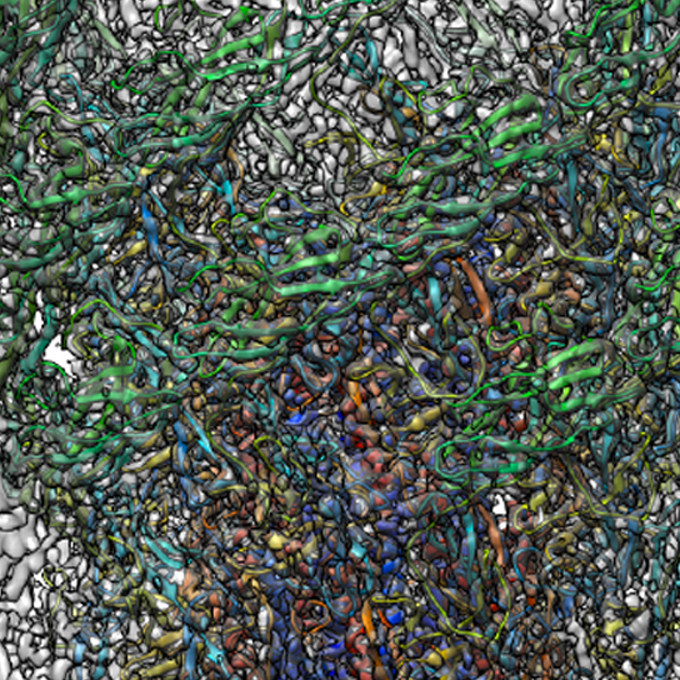
Structure of the native supercoiled flagellar hook as a universal joint
JEOL YOKOGUSHI Research Alliance Laboratories
-
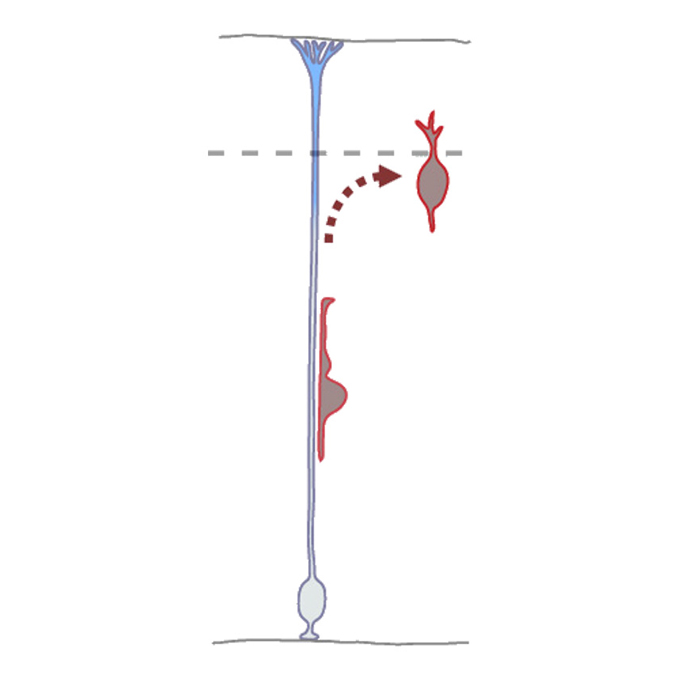
Repulsive signal contributes to proper termination of neuronal migration
Cellular and Molecular Neurobiology Group
-

Identification of a novel arthritis-associated osteoclast precursor macrophage regulated by FoxM1
Laboratory of Immunology and Cell Biology
-
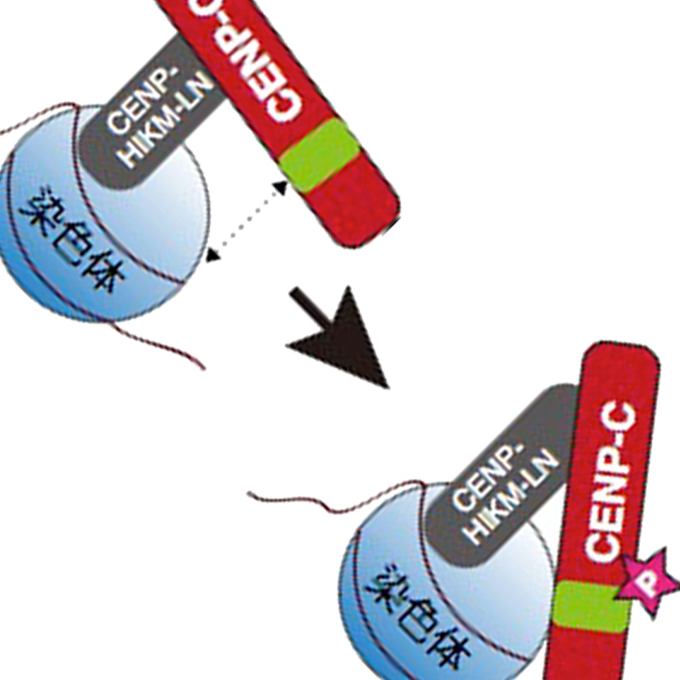
CDK1-mediated CENP-C phosphorylation modulates CENP-A binding and mitotic kinetochore localization
Laboratory of Chromosome Biology
-
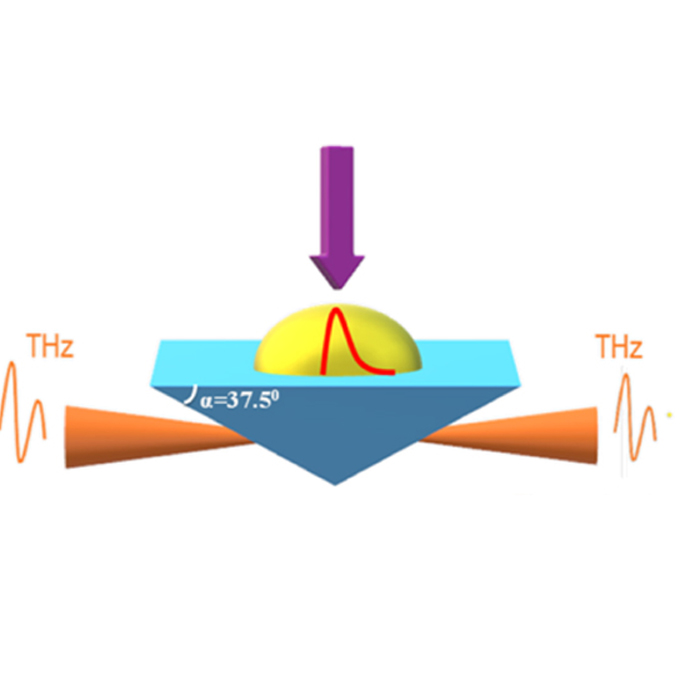
Relaxation dynamics of [Re(CO)2(bpy){P(OEt)3}2](PF6) in TEOA solvent measured by time-resolved attenuated total reflection terahertz spectroscopy
Photophysics Laboratory
-
Epiblast formation by TEAD-YAP-dependent expression of pluripotency factors and competitive elimination of unspecified cells
Laboratory for Embryogenesis
-
Nuclear Formation Induced by DNA-conjugated Beads in Living Fertilised Mouse Egg
Nuclear Dynamics Group
-
Asymmetrical localization of Nup107-160 subcomplex components within the nuclear pore complex in fission yeast
Nuclear Dynamics Group
-
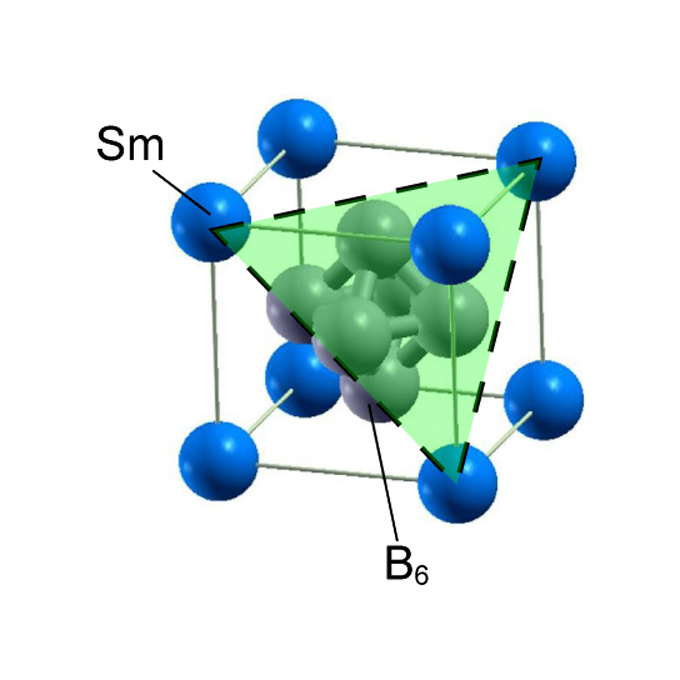
Non-trivial surface states of samarium hexaboride at the (111) surface
Photophysics Laboratory
-
Suppression of autophagic activity by Rubicon is a signature of aging
Laboratory of Intracellular Membrane Dynamics
-
Collective Cell Migration of Dictyostelium Without cAMP Oscillations at Multicellular Stages
Laboratory of Single Molecule Biology
2018
-
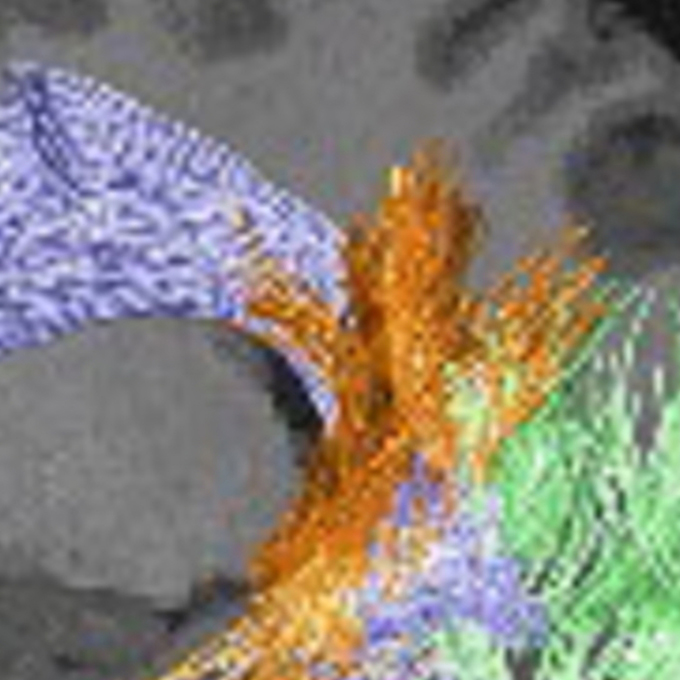
White Matter Pathway and Individual Variability in Human Stereoacuity
Cognitive Neuroscience Group
-
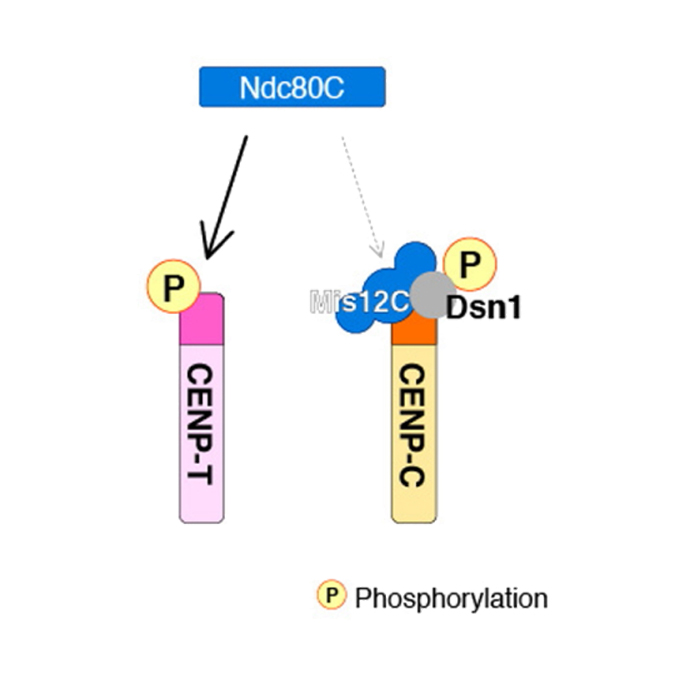
Pulling the Genome Apart: Chromosome Segregation During Mitosis Explained
Laboratory of Chromosome Biology
-
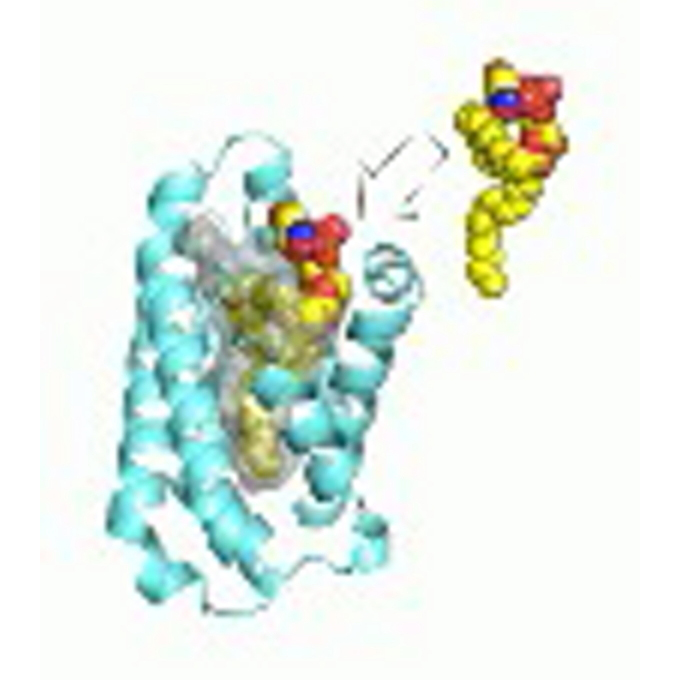
Gip1 structure places G proteins in lockdown
Laboratory of Single Molecule Biology
-

3D genomic architecture reveals that neocentromeres associate with heterochromatin regions
Laboratory of Chromosome Biology
-
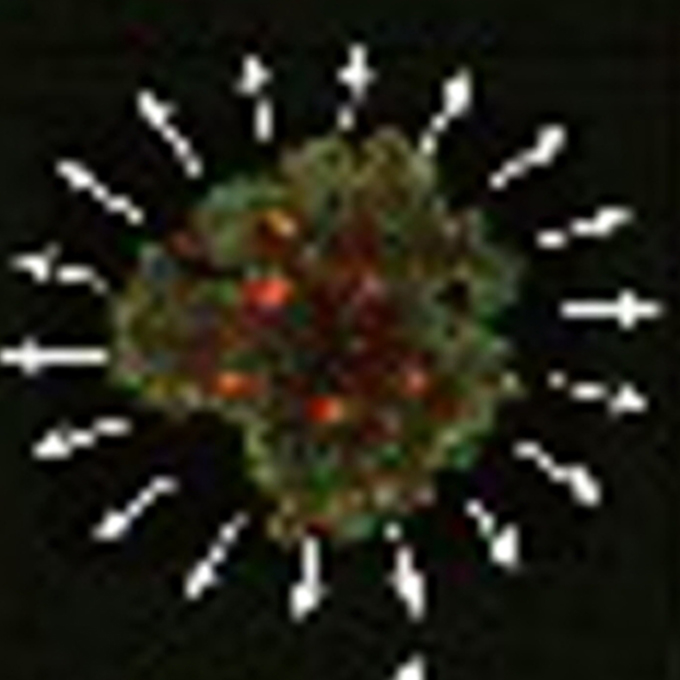
Molecular Inhibition Gets Cells on the Move
Laboratory of Single Molecule Biology
-
Artificial Intelligence Aids Automatic Monitoring of Single Molecules in Cells
Laboratory of Single Molecule Biology
-
Motor error in parietal area 5 and target error in area 7 drive distinctive adaptation in reaching
Dynamic Brain Network Laboratory
-
Insight into structural remodeling of the FlhA ring responsible for bacterial flagellar type III protein export
JEOL YOKOGUSHI Research Alliance Laboratories
-
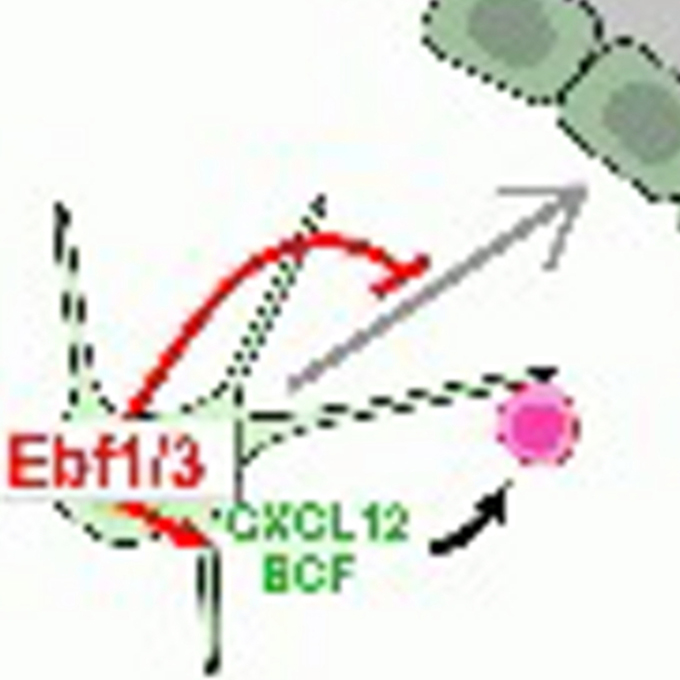
Stem cell niche-specific Ebf3 maintains the bone marrow cavity
Laboratory of Stem Cell Biology and Developmental Immunology
2017
-

Na+-induced structural transition of MotPS for stator assembly of the Bacillus flagellar motor
JEOL YOKOGUSHI Research Alliance Laboratories
-
Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex
JEOL YOKOGUSHI Research Alliance Laboratories
-
Genome Stability by DNA polymerase β in Neural Progenitors Contributes to Neuronal Differentiation in Cortical Development
Cellular and Molecular Neurobiology Group
-
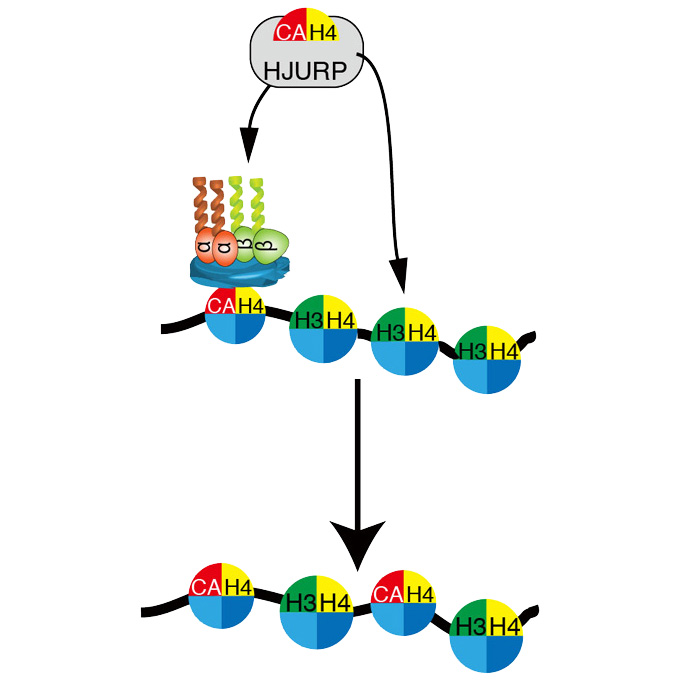
Association of M18BP1/KNL2 with CENP-A nucleosome is essential for centromere formation in non-mammalian vertebrates
Laboratory of Chromosome Biology
-
Endothelial cells are intrinsically defective in xenophagy of Streptococcus pyogenes
Laboratory of Intracellular Membrane Dynamics
-
Brain Alpha rhythm controlling signal traffics in the brain
Dynamic Brain Network Laboratory
-
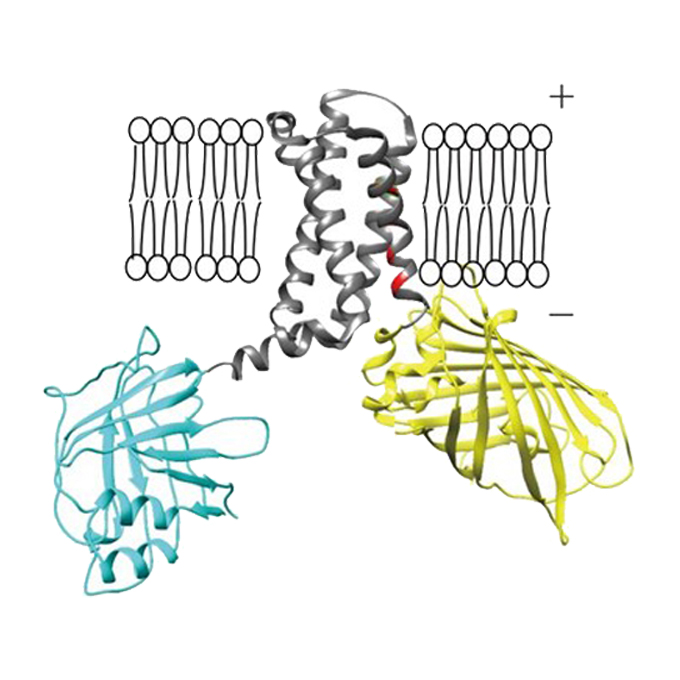
Genetically encoded bioluminescent voltage indicator for multi-purpose use in wide range of bioimaging
-
Identical folds used for distinct mechanical functions of the bacterial flagellar rod and hook
JEOL YOKOGUSHI Research Alliance Laboratories
-
Effects of forced movements on learning: Findings from a choice reaction time task in rats
Cognitive Neuroscience Group
-
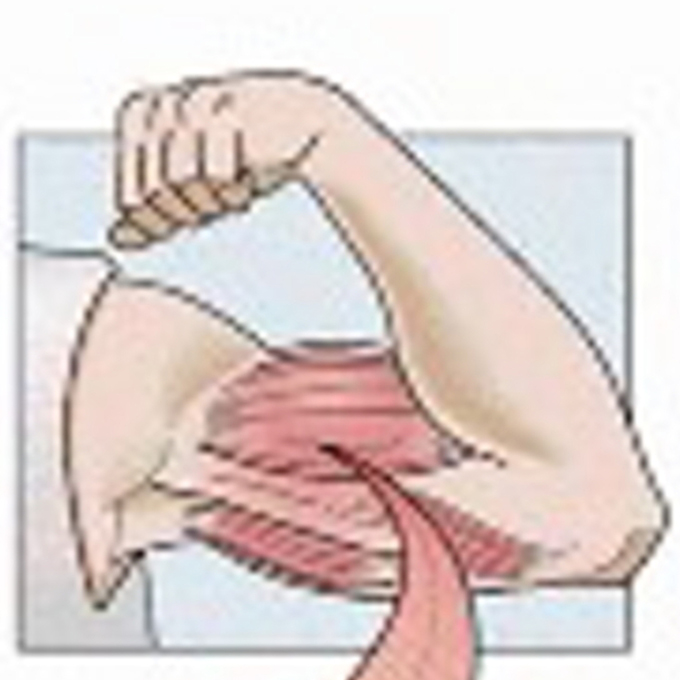
Structure of actomyosin rigour complex at 5.2-Å resolution and insights into the ATPase cycle mechanism
JEOL YOKOGUSHI Research Alliance Laboratories
2016
-
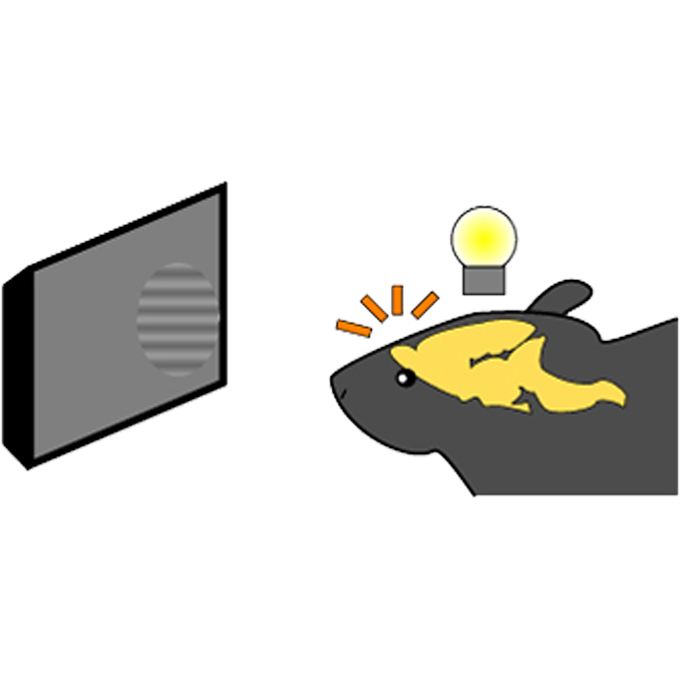
Noradrenaline Improves Behavioral Contrast Sensitivity via the β-Adrenergic Receptor.
-
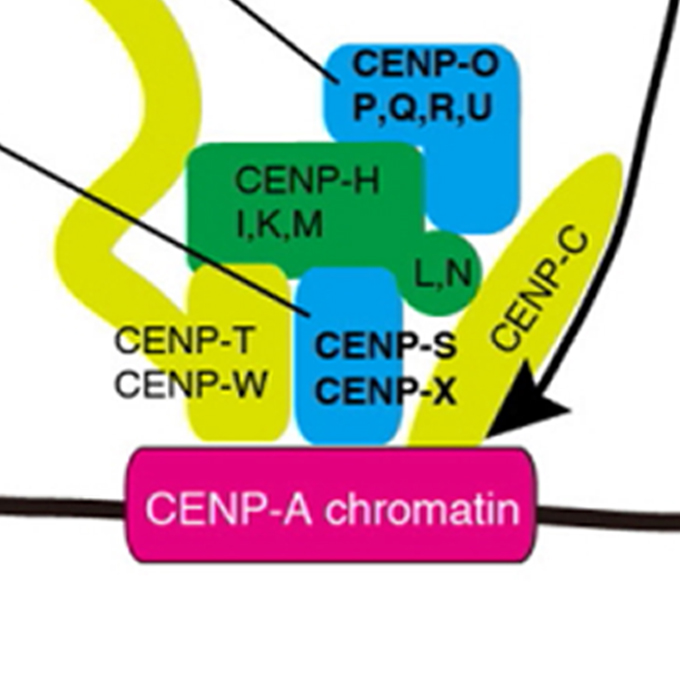
Constitutive centromere-associated network controls centromere drift in vertebrate cells
Laboratory of Chromosome Biology
-
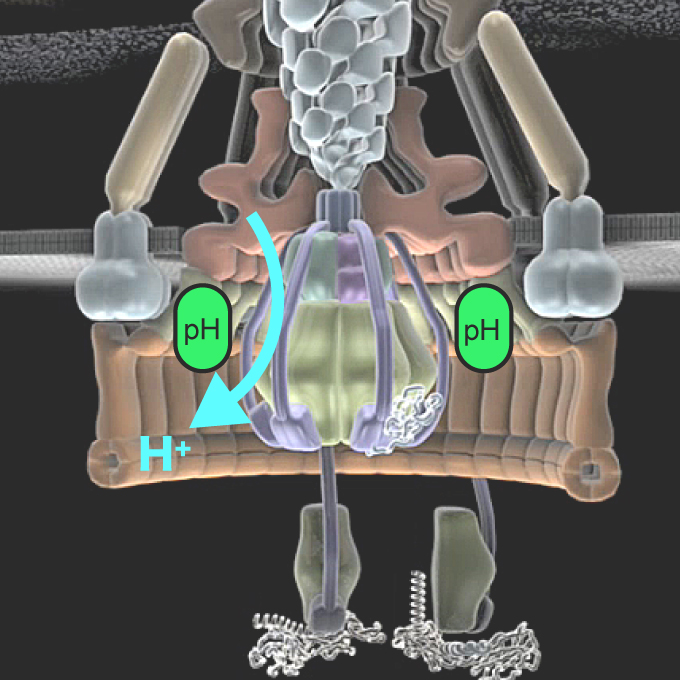
High-resolution pH imaging of living bacterial cell to detect local pH differences
JEOL YOKOGUSHI Research Alliance Laboratories
-
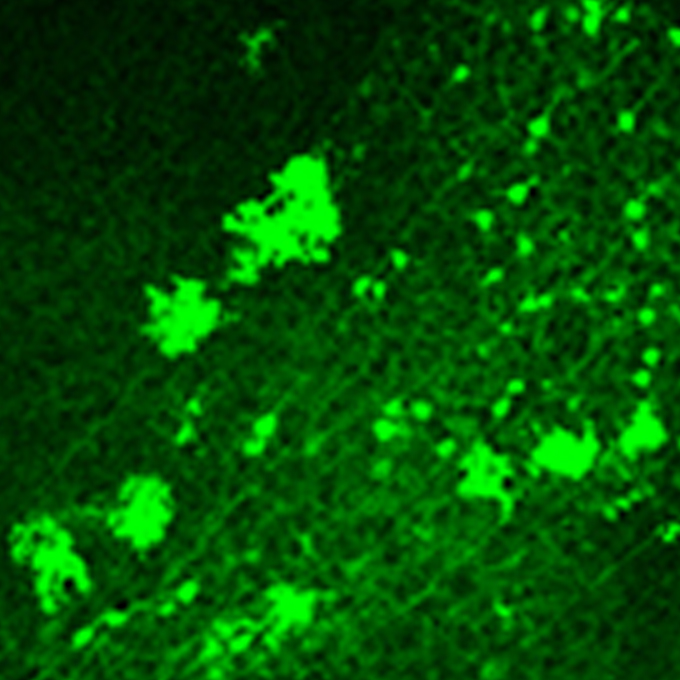
Establishment of high reciprocal connectivity between clonal cortical neurons is regulated by the Dnmt3b DNA methyltransferase and clustered protocadherins
KOKORO-Biology Group
-
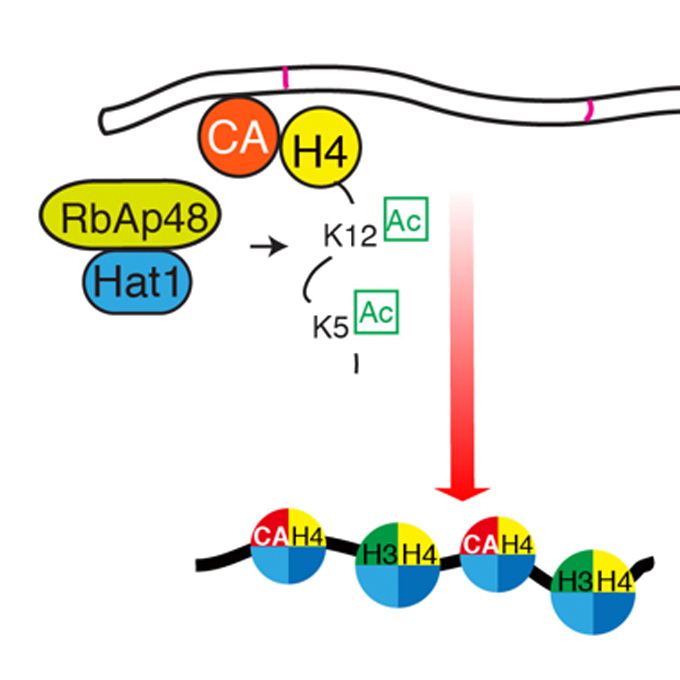
Acetylation of histone H4 Lysine 5 and 12 is required for CENP-A deposition into centromeres
Laboratory of Chromosome Biology
