Maternal iron deficiency causes male-to-female sex reversal in mouse embryos
Researchers from The University of Osaka have revealed the crucial role that iron plays in sex determination of mammalian embryos
| Journal | Nature 643(8070):262-270 (2025) |
|---|---|
| Title | Maternal iron deficiency causes male-to-female sex reversal in mouse embryos |
| Laboratory | Laboratory of Epigenome Dynamics〈Prof. TACHIBANA Makoto〉 |
Developmentally expressed genes play a fundamental role in the formation of a wide range of organs. The Y chromosome-located mammalian sex-determining gene Sry, required for the formation of testis, is activated only within a narrow time window during embryogenesis. However, the processes behind the activation of Sry remain uncertain.
Now, in a recent publication from Nature, a team from The University of Osaka has discovered that iron plays a crucial role in male mammalian sex development.
The Sry gene directs sexually undifferentiated gonads to differentiate into testes rather than ovaries. The Sry gene is initially inactive because of a modification added to DNA-wrapping proteins called histone methylation. In males, a protein called KDM3A is responsible for removing this histone methylation, allowing the Sry gene to become active.
"We already knew that KDM3A requires iron for its enzymatic activity," says lead author, Naoki Okashita. "Therefore, we wanted to investigate the extent and nature of iron metabolism and histone demethylation in sex determination."
The researchers established mice lacking Tfrc, a gene crucial for cellular iron incorporation. Loss of Tfrc prevented the removal of the histone methylation, leading to reduced Sry expression. When inspecting the genitals of these mice, it was found that some XY mice developed as female, despite being genetically male.
The team also established an organ culture system for gonadal cells in which they could manipulate the levels of available iron. This confirmed that iron deficiency reduces the activity of KDM3A and activation of Sry. Therefore, the reduced Sry suppression under iron-deficient conditions was caused by the failure of KDM3A to remove histone methylation.
Perhaps most significantly, they then showed that maternal iron deficiency – whether induced by diet or by pharmacological intervention – could lead to altered histone methylation of the Sry gene because of reduced KDM3A activity. Some population of XY offspring derived from iron-deficient pregnant females were shown to undergo male-to-female sex reversal, potentially due to the impaired Sry activation.
"Our findings indicate that iron deficiency leads to disorders of male development, at least in mice. Our study emphasizes the importance of maintaining maternal iron levels," explains senior author, Makoto Tachibana.
It is known that in humans, Fanconi anemia and Diamond–Blackfan anemia are risk factors for disorders of development. Understanding how iron supports embryonic development underscores the importance of maternal iron intake for the healthy development of human embryos.
Abstract
Ferrous iron (Fe2+) is essential in all eukaryotic cells for various oxidoreductase reactions, including the demethylation of DNA and proteins. We have previously shown that histone demethylation is required for normal epigenetic regulation of the Y-chromosomal sex-determining gene Sry in developing gonads during male sex determination1,2. Here we investigate the potential connection between iron metabolism, histone demethylation and sex determination in mammals. We found that Fe2+-producing pathways are substantially activated in mouse embryonic gonads in the sex-determining period. Chelation of iron in cultured XY gonads reduced the level of KDM3A-mediated H3K9 demethylation of Sry, mostly abolished Sry expression and caused the gonads to express ovarian markers. In vivo, conditional deletion of the gene Tfrc—which is required for iron incorporation—in fetal XY gonadal somatic cells, or acute pharmaceutical suppression of available iron in pregnant mice, resulted in male-to-female gonadal sex reversal in a proportion of the wild-type offspring, highlighting the pivotal role of iron metabolism in male sex determination. Finally, long-term feeding of pregnant mice with a low-iron diet, when combined with a heterozygous variant of Kdm3a that by itself has no observable effect, suppressed Sry expression and caused male-to-female sex reversal in some of the progeny, revealing a connection between maternal dietary iron and fetal developmental outcomes.

Fig. 1
Photograph of offspring born to iron chelator-treated mothers during pregnancy. Left, normally developed XY male animal; middle, intersex-phenotype XY animal; right; completely feminized XY animal.
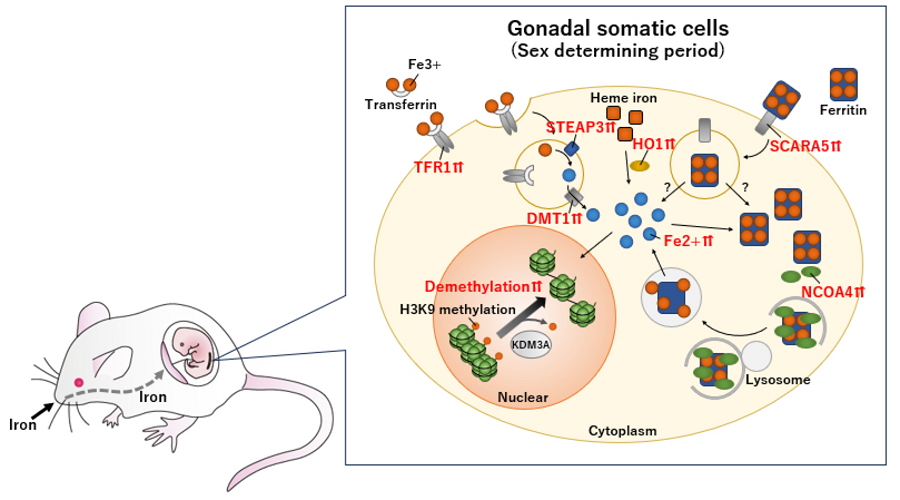
Fig. 2
The role of iron metabolism in mouse male sex determination: intracellular iron elevation drives H3K9 demethylation required for Sry activation.
| Authors | Naoki Okashita (1), Ryo Maeda (1), Shunsuke Kuroki (1), Kyona Sasaki (1), Yoko Uno (1), Peter Koopman (2), Makoto Tachibana (1)
|
|---|---|
| PubMed | 40468068 |
