Biomolecular Networks Laboratories
Laboratory of Epigenome Dynamics
 Prof. TACHIBANA Makoto
Prof. TACHIBANA Makoto
Keywords:
Epigenetics, Histone modification, Development/Differentiation, Reproduction
Understanding the biological significance of epigenome dynamics in mammalian development/differentiation
Our body contains approximately 200 types of cells derived from one fertilized egg. Why do these cells have various functions despite having the same genomic information? One mechanism explaining this issue is epigenetic regulation. Epigenetic regulation represents DNA methylation and histone modification. The modified genome is called "epigenome". Recent research has revealed that epigenome fluctuates dynamically throughout life cycle and closely participates in various life phenomena. Our goal is to clarify the biological significance and molecular mechanisms of epigenome dynamics in mammalian development/differentiation.
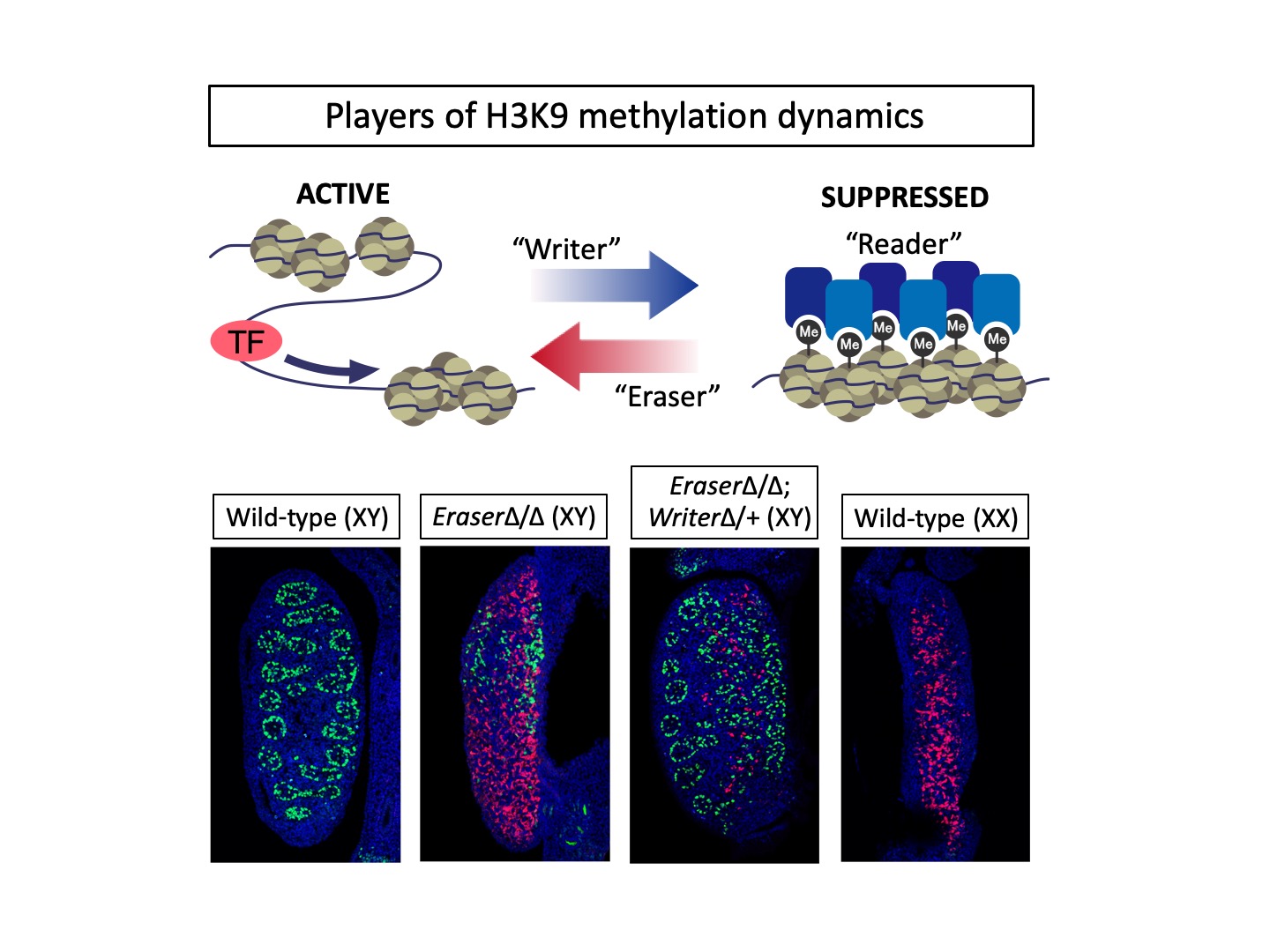
Top: Epigenetic regulation by H3K9 methylation. The three players are involved: methyltransferase as "Writer", demethylase as "Eraser", and modification-specific binding molecule as “Reader”. Mouse embryonic gonads co-stained with antibodies against male cells (green) and female cells (red). Gonadal sex is controlled by the balance between H3K9 methylation and demethylation.
Members
| Makoto Tachibana (Professor) | tachibana.makoto.fbs[at]osaka-u.ac.jp |
|---|---|
| Shunsuke Kuroki (Associate Professor) | kuroki.shunsuke.fbs[at]osaka-u.ac.jp |
| Nanoki Okahita (Assistant Professor) | okaokaoka.fbs[at]osaka-u.ac.jp |
| Ryou Maeda (Specially Appointed Assistant Professor) | rmaeda.fbs[at]osaka-u.ac.jp |
| Tomoko Takeuchi (Secretary) | |
You could probably reach more information of individual researchers by Research Map and researcher's search of Osaka-U.
- ※Change [at] to @
Q&A
- What is your hot research topic?
- We found that epigenetic regulation by histone modification is important for sex determination in mammal. If histone modification is not performed correctly at the certain period of embryogenesis in mice, they cannot become male despite having the Y chromosome. Not only in this case, it has been found that epigenetic gene regulation plays an important role in mammalian reproductive function. We are currently conducting the research to clarify the role of epigenetic regulation in sex determination and gametogenesis.
- What is your breakthrough or research progress in the last 5 years?
- A chromosomal region where gene expression is suppressed is called as heterochromatin. Methylation at lysine 9 of histone H3 (H3K9me) is a representative "mark" of heterochromatin. So far, H3K9me has been considered a very stable modification and will not be removed throughout life. However, we have found that H3K9me fluctulates dynamically in a cell type-specific and temporally specific manner in mammalian life. We clarified that the dynamic change of H3K9me is achieved by the cooperative action of methyltransferase and demethylase, and that the dynamic change of H3K9me is essential for regulating "key" genes of cellular function.
- What kind of background do your lab members have?
- Ph.D staff graduated from agriculture, science and veterinary medicine.
- Do you collaborate with other institutions and universities?
- There are about 40 researchers and advisors who had been involved in PRESTO "Epigenetics". I have been interacting with them for over 10 years.
- How do you develop your research?
- We find that metabolism may play an important role for changing epigenome. Based on this finding, we would like to clarify how metabolic changes alter epigenome during mammalian embryogenesis.
Research Highlights
Publications (Research Articles, Reviews, Books)
2023
CDYL reinforces male gonadal sex determination through epigenetically repressing Wnt4 transcription in mice.
Proc Natl Acad Sci USA 120(20):e2221499120. 2023 (PMID:37155872 DOI:10.1073/pnas.2221499120.)
2022
Spectrum of Sex : The Molecular Bases that Induce Various Sexual Phenotypes
Springer ISBN: 978-981-19-5359-0 2022
HP1 maintains protein stability of H3K9 methyltransferases and demethylases.
EMBO Rep e53581 2022 (PMID:35166421 DOI:10.15252/embr.202153581)
2021
Meiosis-specific ZFP541 repressor complex promotes developmental progression of meiotic prophase towards completion during mouse spermatogenesis.
Nat Commun 12(1):3184 2021 (PMID:34075040 DOI:10.1038/s41467-021-23378-4)
Generation of ovarian follicles from mouse pluripotent stem cells.
Science 373(6552):eabe0237 2021 (PMID:34437124 DOI:10.1126/science.abe0237)
Dynamic erectile responses of a novel penile organ model utilizing TPEM.
Biol Reprod 104(4):875-886 2021 (PMID:33511393 DOI:10.1093/biolre/ioab011)
2020
The mouse Sry locus harbors a cryptic exon that is essential for male sex determination
Science 370(6512):121-124 2020 (PMID:33004521 DOI:10.1126/science.abb6430)
The PRDM14-CtBP1/2-PRC2 complex regulates transcriptional repression during the transition from primed to naïve pluripotency
Journal of Cell Science 133(15) 2020 (PMID:32661086 DOI:10.1242/jcs.240176 )
H3K9 demethylases JMJD1A and JMJD1B control prospermatogonia to spermatogonia transition in mouse germline
Stem Cell Reports 15(2):424-438 2020 (PMID:32679061 DOI:10.1016/j.stemcr.2020.06.013 )
G9a is involved in the regulation of critical bone formation through activation of Runx2 function during development
Bone 137:115332 2020 (PMID:32344102 DOI:10.1016/j.bone.2020.115332 )
Caspase-8, receptor-interacting protein kinase 1 (RIPK1), and RIPK3 regulate retinoic acid-induced cell differentiation and necroptosis.
Cell Death Differ. 27(5):1539-1553 2020 (PMID:31659279 DOI:10.1038/s41418-019-0434-2 )
2019
Interferon-γ induces the cell surface exposure of phosphatidylserine by activating the protein MLKL in the absence of caspase-8 activity.
J. Biol. Chem. 294(32):11994-12006 2019 (PMID:31217278 DOI:10.1074/jbc.RA118.007161)
TET2 catalyzes active DNA demethylation of the Sry promoter and enhances its expression.
Sci Rep 9(1):13462 2019 (PMID:31530896 DOI:10.1038/s41598-019-50058-7)
Role of epigenetic regulation in mammalian sex determination.
Curr. Top. Dev. Biol. 134:195-221 2019 (PMID:30999976 DOI:10.1016/bs.ctdb.2019.01.008)
Histone H3K9 Methyltransferase G9a in Oocytes Is Essential for Preimplantation Development but Dispensable for CG Methylation Protection.
Cell Reports 27(1):282-293.e4 2019 (PMID:30943408 DOI:10.1016/j.celrep.2019.03.002)
G9a-dependent histone methylation can be induced in G1 phase of cell cycle.
Sci Rep 9(1):956 2019 (PMID:30700744 DOI:10.1038/s41598-018-37507-5)
2018
DNMTs and SETDB1 function as co-repressors in MAX-mediated repression of germ cell-related genes in mouse embryonic stem cells.
PLoS One 13(11):e0205969 2018 (PMID:30403691 DOI:10.1371/journal.pone.0205969)
FGF2 Has Distinct Molecular Functions from GDNF in the Mouse Germline Niche.
Stem Cell Rep. 10(6):1782-1792 2018 (PMID:29681540 DOI:10.1016/j.stemcr.2018.03.016)
Combined Loss of JMJD1A and JMJD1B Reveals Critical Roles for H3K9 Demethylation in the Maintenance of Embryonic Stem Cells and Early Embryogenesis.
Stem Cell Rep. 10(4):1340-1354 2018 (PMID:29526734 DOI:10.1016/j.stemcr.2018.02.002)
Epigenetic regulation of mammalian sex determination.
Mol. Cell. Endocrinol. 468:31-38 2018 (PMID:29248548 DOI:10.1016/j.mce.2017.12.006)
Our ideal candidate (as a graduate student)
We are looking for a highly motivated person to work on our research topics as our lab member. Our lab welcomes the person who loves taking care of creatures, hand working and handcraft too. Any kind of background (such as your expertise or major) is available.
Contact
Laboratory of Epigenome Dynamics, Graduate School of Frontier Biosciences, Osaka University,
1-3 Yamadaoka, Suita, Osaka 565-0871 Japan.
TEL:+81-6-6879-4670
E-mail: tachibana.makoto.fbs[at]osaka-u.ac.jp (Prof. Makoto Tachibana)
- ※Change [at] to @
