Molecular Inhibition Gets Cells on the Move
| Journal | Nat Commun 9, 4481 (2018) |
|---|---|
| Title | Molecular Inhibition Gets Cells on the Move |
| Laboratory | Laboratory of Single Molecule Biology〈Prof. UEDA Masahiro〉 |
To respond to changes in their environment, individual cells, including some of those that compose complex organisms, need to move. This can be accomplished by propelling themselves using projections called pseudopodia. Although this system is present within many different species, the mechanisms that control it and enable cells to move in a particular direction have not been completely clarified.
In recent work published in the journal Nature Communications, researchers at Osaka University have revealed how cells in an organism known as a slime mold establish the polarized distribution of two molecules in their outer membranes. The localization of these two molecules exclusively at different ends of a cell leads to the assembly of machinery for pseudopodium formation only at one cell end. This ensures a unidirectional force from pseudopodia used to propel cells, enabling directed cell movement.
The researchers used a number of single-cell and single-molecule experimental approaches to analyze the PIP3 and PTEN molecules in slime mold and determine how their polarized distribution arises. First, they showed that when PTEN was absent from the cells, PIP3 became distributed across the whole of the cell membrane, which led to multiple pseudopodia being generated. This in turn prevented cell movement. They also quantified the levels of PIP3 and PTEN, and their specific cellular distributions, and showed that they were exclusively distributed in different regions on the membrane, with a clear boundary between them.
"Our findings reveal that PTEN and PIP3 function as an ultrasensitive switch in cells," says author Masahiro Ueda. "The presence of PTEN and PIP3 means they mutually suppress each other, which prevents cells from forming pseudopodia at different ends. This is an extremely effective way of guaranteeing that cells will generate propulsive forces in only one direction, avoiding wasted energy."
Given that PTEN and PIP3 appear to exert similar functions in a range of organisms from slime mold up to mammals, these findings could explain cell motility in many species. The setup of having two positive feedback loops of mutually inhibitory molecules may also function in other signaling pathways, given the efficiency with which it enables ultrasensitive switching between different states.
"Our work also hints at how this system might function to promote cell viability," says lead author Satomi Matsuoka. "For example, when a chemoattract is present with a particular concentration gradient, it causes the PIP3-rich region in the cell membrane to orient itself relative to that gradient, which in turn induces propulsion in the direction of the gradient. In this way, cells are automatically induced to move towards or away from stimuli and toxic chemicals in their environment."
Abstract
Phosphatidylinositol 3,4,5-trisphosphate (PIP3) and PIP3 phosphatase (PTEN) are enriched mutually exclusively on the anterior and posterior membranes of eukaryotic motile cells. However, the mechanism that causes this spatial separation between the two molecules is unknown. Here we develop a method to manipulate PIP3 levels in living cells and used it to show PIP3 suppresses the membrane localization of PTEN. Single-molecule measurements of membrane-association and -dissociation kinetics and of lateral diffusion reveal that PIP3 suppresses the PTEN binding site required for stable PTEN membrane binding. Mutual inhibition between PIP3 and PTEN provides a mechanistic basis for bistability that creates a PIP3 -enriched/PTEN-excluded state and a PTEN-enriched/PIP3 -excluded state underlying the strict spatial separation between PIP3 and PTEN. The PTEN binding site also mediates the suppression of PTEN membrane localization in chemotactic signaling. These results illustrate that the PIP3-PTEN bistable system underlies a cell's decision-making for directional movement irrespective of the environment.
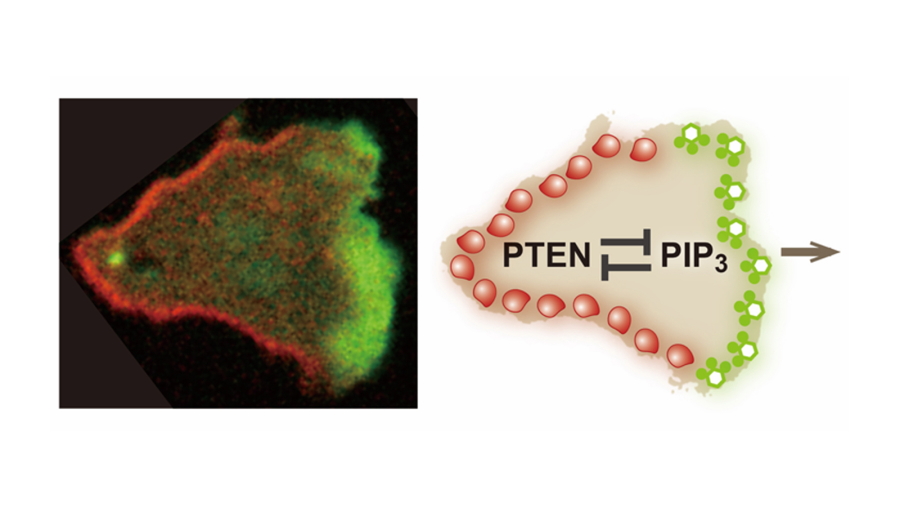
Fig. 1
Mutually exclusive distribution of PIP3 and PTEN on the plasma membrane of a migrating cell.
(left) A Dictyostelium discoideum cell migrating rightward. The cell expresses PIP3-binding protein tagged with green fluorescent protein (GFP) and PTEN labeled with tetramethylrhodamine (TMR). A PIP3-enriched domain and a PTEN-enriched domain emerge at the front and back of the migrating cell in a mutually exclusive manner. The major question examined in this study is how the PIP3-enriched domain and the PTEN-enriched domain are clearly separated. (right) An illustration summarizing the main point of this study. This study reveals mutual inhibition between PTEN and PIP3, which provides the mechanistic basis for the clear spatial separation between the PIP3-enriched domain and the PTEN-enriched domain.
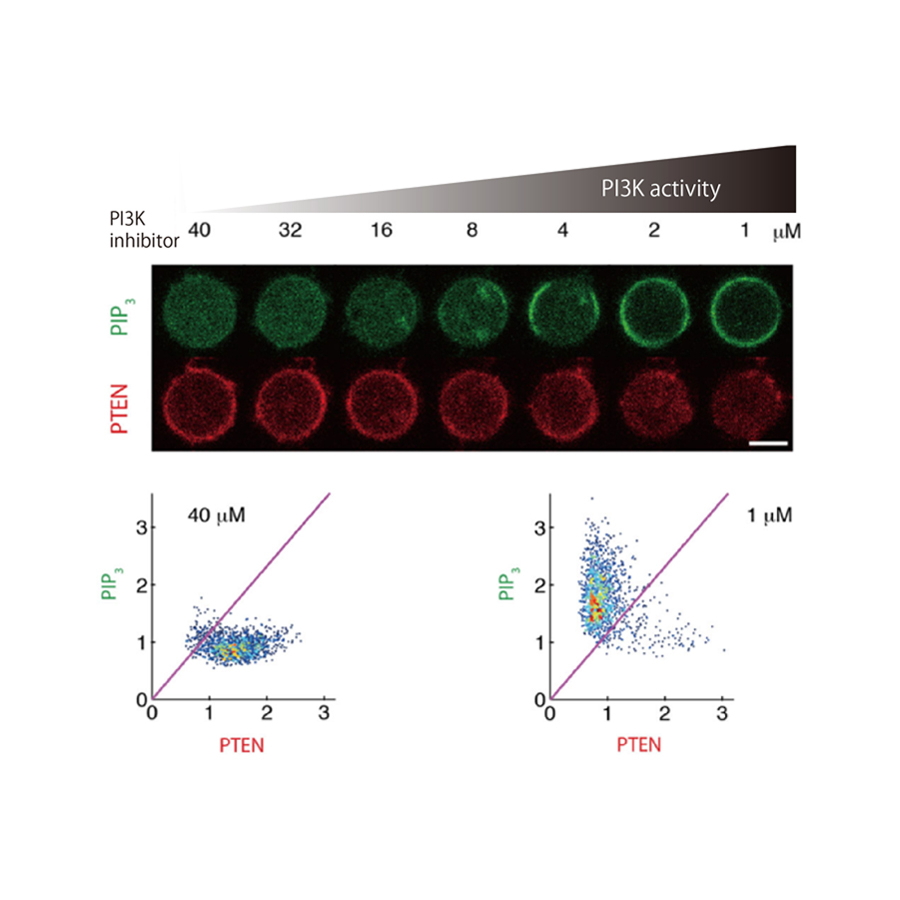
Fig. 2
Mutual inhibition between PIP3 and PTEN.
The reciprocal enrichment of PIP3 and PTEN on the plasma membrane of Dictyostelium discoideum cells upon a gradual decrease in the inhibitor concentration against PI3K, a PIP3-producing enzyme. (upper) Under low-PI3K-activity conditions, PIP3 was not enriched on the plasma membrane. Under moderate-activity conditions, PIP3 abruptly became enriched in response to a slight increase in PI3K activity. Under the high-PI3K-activity conditions, PIP3 was enriched on the membrane. PTEN exhibited a mirrored response to PIP3 toward the gradual increase in PI3K activity. (lower) PIP3-depleted/PTEN-enriched state was adopted over almost all of the membrane under the lowest-activity conditions, while the state was changed to PIP3-enriched/PTEN-depleted state everywhere under the highest conditions.
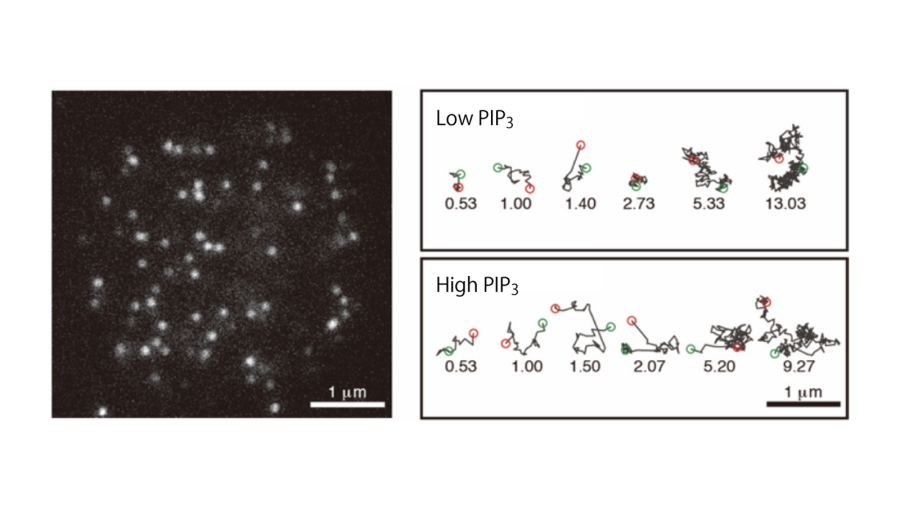
Fig. 3
Single-molecule imaging analysis of the interaction between PTEN and the plasma membrane.
(left) A total internal reflection fluorescence microscopy (TIRFM) image of the living Dictyostelium discoideum cell expressing PTEN labeled with TMR. White spots represent single PTEN molecules bound to the plasma membrane at the bottom of the cell. (right) Trajectories of representative single PTEN molecules on the membrane. When bound to the high-PIP3 membrane, PTEN exhibited a shorter lifetime of membrane interaction and faster lateral diffusion than when bound to the low-PIP3 membrane.
| Authors | Matsuoka S (1, 2, 3), Ueda M (4, 5, 6, 7)
|
|---|---|
| PubMed | 30367048 |
