Nanobiology Laboratories
Laboratory of Single Molecule Biology
 Prof. UEDA Masahiro
Prof. UEDA Masahiro
Keywords:
Single-molecule imaging, Mathematical modeling, Dictyostelium discoideum, Signal transduction, Fluctuation
Understanding molecular stochastic computation in intracellular signaling systems
We are interested in cellular functions such as intracellular signaling and cellular motility. In particular, we focus on questions such as “how do these cellular properties arise from reaction networks composed of stochastically-operating biomolecules?” and “what are the mechanisms that enable the intracellular signaling system to be robust to molecular noise, and sometimes utilize that noise to express its functions?”. Our research group develops and utilizes cutting-edge measurement techniques such as single-molecule imaging and mathematical modeling. We aim to understand the design principles underlying the remarkable signal processing capability of living cells with single-molecule resolution.
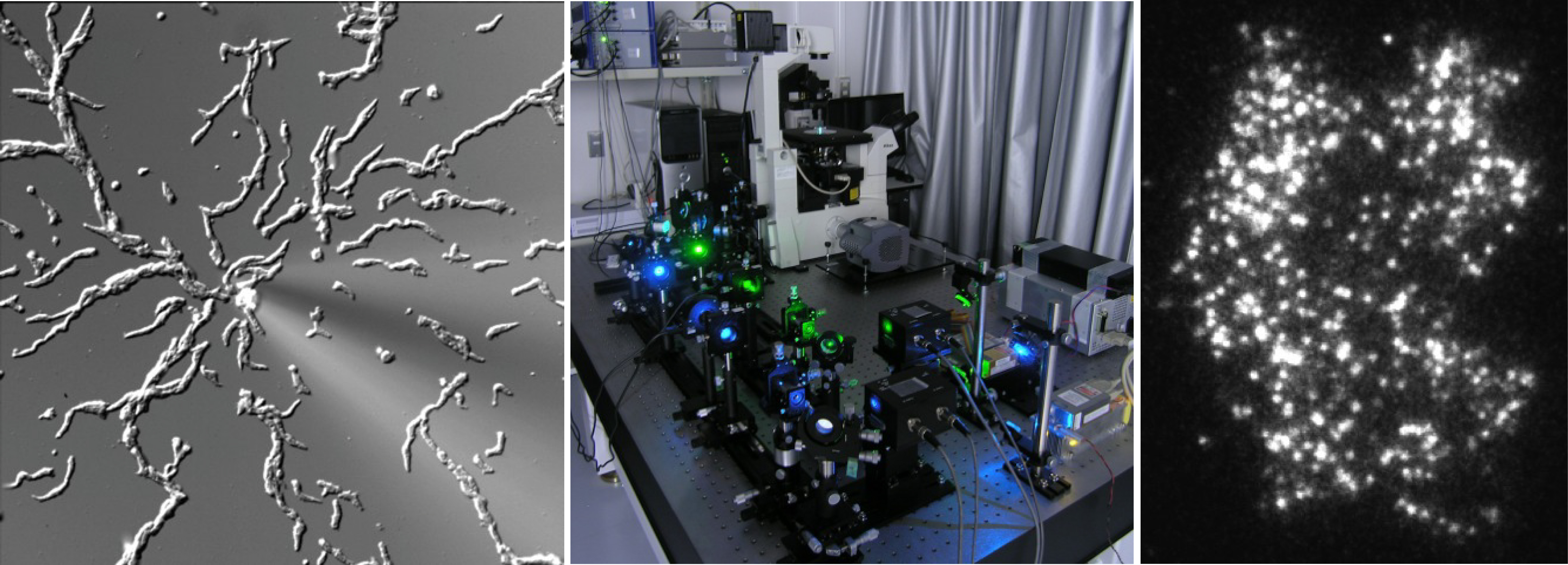
(Left) Chemotaxis of the cellular slime mold Dictyostelium discoideum. (Middle) TIRFM for single-molecule imaging in living cells. (Right) Single-molecules of PTEN.
Members
| UEDA Masahiro (Professor) | ueda.masahiro.fbs[at]osaka-u.ac.jp |
|---|---|
| HIROSHIMA Michio (Specially Appointed Professor) | m_hiroshima.fbs[at]osaka-u.ac.jp |
| ARIGA Takayuki ( Associate Professor) | ariga.fbs[at]osaka-u.ac.jp |
| MATSUOKA Satomi (Assistant Professor) | matsuoka.satomi.fbs[at]osaka.u.ac.jp |
| MIURA Kota (guest Professor) | kota.miura.fbs[at]osaka-u.ac.jp |
You could probably reach more information of individual researchers by Research Map and researcher's search of Osaka-U.
- ※Change [at] to @
Q&A
- What is your hot research topic?
- We are interested in cellular functions, such as intracellular information processing and cell motility. In particular, we focus on questions such as "how do these cellular properties spontaneously arise from biomolecules and reaction networks composed of biomolecules?", and "what are the mechanisms that enable the information processing system to be robust to intrinsic noise, and sometimes utilize that noise to express its functions while also receiving severe stochastic fluctuations from the external environment?" As a typical example of such a stochastic calculation process, we focus on concentration gradient sensing and the taxis of Dictyostelium discoideum. Our research group utilizes cutting-edge measurement techniques, such as single-molecule imaging; theoretical approaches, including mathematical modeling with the aid of high-performance computers; and in vitro reconstitutions of minimal molecular reaction systems. We aim to understand the design principles underlying the remarkable information processing capability of cells.
- What is your breakthrough or research progress in the last 5 years?
- To understand how molecules act in cells, it would be ideal to be able to track individual molecules, including where in the cell they are located and what modifications they undergo when conditions in the cell change. However, this ideal situation is difficult to actualize with existing technologies, and a huge amount of time would be required to perform such sensitive monitoring. Recently, we developed a system that can overcome these difficulties by automatically searching for, focusing on, imaging, and tracking single molecules within living cells. Using this system, we were able to analyze hundreds of thousands of single molecules in hundreds of cells in a short period, thereby providing reliable data about the status and dynamics of molecules of interest.
- What kind of background do your lab members have?
- Biology, engineering, physics, chemistry, mathematics, information science, and so on.
- Do you collaborate with other institutions and universities?
- We have been collaborating with the RIKEN Center for Life Sciences since 2011. Overseas, we have collaborated with Peter N. Devreotes, Miho Iijima (Johns Hopkins University), Tian Jin (NIH), Peter van Haastert (University of Groningen) and others.
The research includes imaging analysis of intracellular signal transduction systems and development of single-molecule imaging methods.
- What kind of careers do your Lab's alumni go on to?
- Academia researchers, pharmaceutical company researchers, venture company entrepreneurs, and others.
- How do you develop your research?
- Since our research is extremely basic, it is difficult to foresee. Hopefully, we would like to be able to understand cellular function mechanisms as the spatiotemporal dynamics of the interaction of individual biomolecules.
Research Highlights
Publications (Research Articles, Reviews, Books)_
2025
Tandem electrostatic lens as a zoom system to achieve ultra-high magnification in microscope imaging mass spectrometry
Review of Scientific Instruments Rev. Sci. Instrum. 96, 103301 (2025) 2025 (PMID:41036972 DOI:10.1063/5.0283704)
Excitable Ras dynamics-based screens reveal RasGEFX is required for macropinocytosis and random cell migration
Nature Communications 2;16(1):117. 2025 (PMID:39746985 DOI:0.1038/s41467-024-55389-2.)
2024
Single molecule tracking based drug screening
Nature Communications 17;15(1):8975. 2024 (PMID:39420015 DOI:10.1038/s41467-024-53432-w.)
Site-specific clustering of bioactive signaling molecules predicted in situ by space and time coherent mapping for imaging mass spectrometry
Journal of the American Society for Mass Spectrometry 1;36(1):72-84. 2024 (PMID:39580810 DOI:10.1021/jasms.4c00333.)
Application of single-molecule analysis to singularity phenomenon of cells
Biophysics and Physicobiology 8;21(Supplemental):e211018. 2024 (PMID:39175861 DOI:10.2142/biophysico.bppb-v21.s018)
Automated single-molecule imaging for drug discovery
Proc. SPIE 12853 High-Speed Biomedical Imaging and Spectroscopy IX 128530 2024 ( DOI:10.1117/12.3009718)
Spontaneous Signal Generation by an Excitable System for Cell Migration
Frontiers in Cell and Developmental Biology 28:12:1373609 2024 (PMID:38481533 DOI:10.3389/fcell.2024.1373609.)
Left-right Myosin-Is, Myosin1C, and Myosin1D exhibit distinct single molecule behaviors on the plasma membrane of Drosophila macrophages
Genes to Cells 29(5):380-396. 2024 (PMID:38454557 DOI:10.1111/gtc.13110.)
Cholesterol suppresses spontaneous activation of EGFR-mediated signal transduction
BBRC 16:704:149673 2024 (PMID:38401305 DOI:0.1016/j.bbrc.2024.149673.)
2023
_A dynamic partitioning mechanism polarizes membrane protein distribution
Nature Communications 30;14(1):7909. 2023 (PMID:38036511 DOI:10.1038/s41467-023-43615-2.)
Sphingomyelin metabolism underlies Ras excitability for efficient cell
Cell Structure and Function 31;48(2):145-160. 2023 (PMID:37438131 DOI: 10.1247/csf.23045.)
_migration and chemotaxis
Scientific Report 13(1):6175. 2023 (PMID:37061516 DOI:10.1038/s41598-023-28572-6.)
Field model for multistate lateral diffusion of various transmembrane proteins observed in living Dictyostelium cells
Journal of Cell Science 136 (4): jcs260280. 2023 (PMID:36655427 DOI: 10.1242/jcs.260280)
2022
Calcium responses to external mechanical stimuli in the multicellular stage of Dictyostelium discoideum
Scientific Report 12(1):12428 2022 (PMID:35859163 DOI:10.1038/s41598-022-16774-3)
SRC kinase activator CDCP1 promotes hepatocyte growth factor-induced cellmigration/invasion of a subset of breast cancer cells
J. Biol. Chem. 298, 101630, 2022 (PMID:35085554 DOI:10.1016/j.jbc.2022.101630)
Heterotrimeric Gq act as a switch for GRK5/6 selectivity underlying_β-arrestin transducer bias_
Nature Communications 13(1):487. 2022 (PMID:35078997 DOI:10.1038/s41467-022-28056-7)
2021
Comparative analysis of single-molecule dynamics of TRPV1 and TRPV4 channels in living cells
international Journal of Molecular Sciences 22(16):8473 2021 (PMID:34445178 DOI:10.3390/ijms22168473)
A sub-population of Dictyostelium discoideum cells shows extremely high sensitivity to cAMP for directional migration
Biochem.Biophys.Ras.Commun 554:131-137 2021 (PMID:33784508 DOI:10.1016/j.bbrc.2021.03.095)
Different heterotrimeric G protein dynamics for wide-range chemotaxis in eukaryotic cells In Dictyostelium: A Tractable Cell and Developmental Model in Biomedical Research. (eds. Robin Williams and Robert Huber and Annette M_ller)
Frontiers in Cell and Developmental Biology 2021 (PMID:34414196 DOI:10.3389/fcell.2021.724797)
GPCR signaling regulation in Dictyostelium chemotaxis
Methods in Molecular Biology 2274:317-336 2021
2020
Single-molecule imaging of PI(4,5)P 2 and PTEN in vitro reveals a positive feedback mechanism for PTEN membrane binding
Communications Biology 3(1):92 2020 (PMID:32111929 DOI:10.1038/s42003-020-0818-3)
Intracellular ATP levels influence cell fates in Dictyostelium discoideum differentiation
Genes Cells 25(5):312-326 2020 (PMID:32125743 DOI:10.1111/gtc.12763)
Talin B regulates collective cell migration via PI3K signaling in Dictyostelium discoideum mounds
Biochem. Biophys. Res. Commun. 525(2):372-377 2020 (PMID:32098673 DOI:10.1016/j.bbrc.2020.02.060)
Large scale single-molecule imaging aided by artificial intelligence
Microscopy 69(2):69-78 2020 (PMID:32090254 DOI:10.1093/jmicro/dfz116)
2019
Transducin activates cGMP phosphodiesterase by trapping inhibitory γ subunit freed reversibly from the catalytic subunit in solution.
Sci Rep 5.40625 2019 (PMID:31076603 DOI:10.1038/s41598-019-43675-9)
Excitable dynamics of Ras triggers spontaneous symmetry breaking of PIP3 signaling in motile cells
J. Cell Sci. 132:jcs.224121 2019 (PMID:30745337 DOI:10.1242/jcs.224121)
Collective cell migration of Dictyostelium without cAMP oscillations at multicellular stages
Communications Biology 2:34 2019 (PMID:30701199 DOI:10.1038/s42003-018-0273-6)
2018
Chemoattractant receptors activate, recruit and capture G proteins for wide range chemotaxis
Biochem. Biophys. Res. Commun. 507:304-310 2018 (PMID:30454895 DOI:10.1016/j.bbrc.2018.11.029)
Parallel signaling pathways regulate excitable dynamics differently to mediate pseudopod formation during eukaryotic chemotaxis
J. Cell Sci. 131:jcs214775 2018 (PMID:30404836 DOI:10.1242/jcs.214775)
Structural basis of Gip1 for cytosolic sequestration of G-protein in wide range chemotaxis
Nat. Commun. in press 2018 (PMID:30401901 DOI:10.1038/s41467-018-07035-x)
Mutual inhibition between PTEN and PIP3 generates bistability for polarity in motile cells
Nat. Commun. 3.486805556 2018 (PMID:30367048 DOI:10.1038/s41467-018-06856-0)
Single-molecule diffusion-based estimation of ligand effects on G protein-coupled receptors
Sci. Signal. 11(548):eaao1917 2018 (PMID:30228224 DOI:10.1126/scisignal.aao1917)
Automated single-molecule imaging in living cells
Nat. Commun. 2.500694444 2018 (PMID:30076305 DOI:10.1038/s41467-018-05524-7)
A study of wound repair in Dictyostelium cells by using novel laserporation
Sci Rep 8(1):7969 2018 (PMID:29789591 DOI:10.1038/s41598-018-26337-0)
Transient acceleration of epidermal growth factor receptor dynamics produces higher order signaling clusters
J. Mol. Biol. 430:1381-1396 2018 (PMID:29505756 DOI:10.1016/j.jmb.2018.02.018)
2017
Intracellular protein-labeling probes for multicolor single-molecule imaging of immune receptor-adaptor molecular dynamics
J. Am. Chem. Soc. 139:17397-17404 2017 (PMID:29119782 DOI:10.1021/jacs.7b08262)
2016
Multi-State Transition Kinetics of Intracellular Signaling Molecules by Single-Molecule Imaging Analysis
Methods in Molecular Biology 1407:361-379 2016 (PMID:27271914 DOI:10.1007/978-1-4939-3480-5_25)
Heterotrimeric G-protein shuttling via Gip1 extends the dynamic range of eukaryotic chemotaxis.
Proc. Natl. Acad. Sci. U. S. A. 113:4356-4361 2016 (PMID:27044073 DOI:10.1073/pnas.1516767113)
Our ideal candidate (as a graduate student)
We are looking for a highly motivated person to work on our research topics as our lab member. Our lab welcomes the person who loves taking care of creatures, hand working and handcraft too. Any kind of background (such as your expertise or major) is available.
Contact
Laboratory of Single Molecule Biology, Graduate School of Frontier Biosciences, Osaka University,
1-3 Yamadaoka, Suita, Osaka 565-0871 Japan.
TEL: +81-6-6879-4611
E-mail: masahiroueda[at]fbs.osaka-u.ac.jp (Prof. Masahiro Ueda)
- ※Change [at] to @
