Gip1 structure places G proteins in lockdown
| Journal | Nat Commun 9, 4635 (2018) |
|---|---|
| Title | Gip1 structure places G proteins in lockdown |
| Laboratory | Laboratory of Single Molecule Biology〈Prof. UEDA Masahiro〉 |
Heterotrimeric G proteins are important in G protein-coupled receptor signaling, which plays many roles in the detection of various environmental stimuli, including hormones, neurotransmitters, light, smells, and chemical signals. G protein functions are regulated by interactions with Gip1, a protein that sequesters G proteins to block signaling processes. Many studies have attempted to understand the mechanism for this interaction between G proteins and Gip1; none have provided a clear explanation, until now.
In a new study published in Nature Communications, a research team, led by experts from Osaka University, investigated the structure of Gip1 to better understand how Gip1 sequesters G proteins in the cytosol, away from their primary location at the cell membrane. They found that Gip1 contains a hydrophobic (water-fearing) space, with a unique molecular bonding arrangement that allows interactions with G proteins.
"We found that the Gip1 hydrophobic cavity, as well as the hydrogen bond network at its entrance, are critical in the ability of Gip1 to mask the membrane-binding region of heterotrimeric G proteins," says corresponding author Masahiro Ueda. "Experimentally modifying either of these aspects greatly impacted the abilities of Gip1 to bind G proteins and release those proteins when needed."
In the study, the hydrophobic cavity was shown to involve six alpha helices, which is an unusual structural composition for this type of cavity. Because of this unique structure, although Gip1 is similar to other proteins that bind hydrophobic groups, it solely binds to heterotrimeric G proteins.
"Our analyses revealed unique aspects of solubilization of lipophilic G proteins as a regulatory mechanism during eukaryotic chemotaxis, which we used as a model system," says lead author Takero Miyagawa. "Because heterotrimeric G proteins are involved in many different signal transduction processes, our findings are important to both basic science advancements and potential drug development."
In addition to providing a better understanding of the mechanism for Gip1 interaction with G proteins, the researchers showed how this interaction might be disrupted, which may be important in understanding diseases caused by defects in signal transduction.
Abstract
G protein interacting protein 1 (Gip1) binds and sequesters heterotrimeric G proteins in the cytosolic pool, thus regulating G protein-coupled receptor (GPCR) signalling for eukaryotic chemotaxis. Here, we report the underlying structural basis of Gip1 function. The crystal structure reveals that the region of Gip1 that binds to the G protein has a cylinder-like fold with a central hydrophobic cavity composed of six α-helices. Mutagenesis and biochemical analyses indicate that the hydrophobic cavity and the hydrogen bond network at the entrance of the cavity are essential for complex formation with the geranylgeranyl modification on the Gγ subunit. Mutations of the cavity impair G protein sequestration and translocation to the membrane from the cytosol upon receptor stimulation, leading to defects in chemotaxis at higher chemoattractant concentrations. These results demonstrate that the Gip1-dependent regulation of G protein shuttling ensures wide-range gradient sensing in eukaryotic chemotaxis.
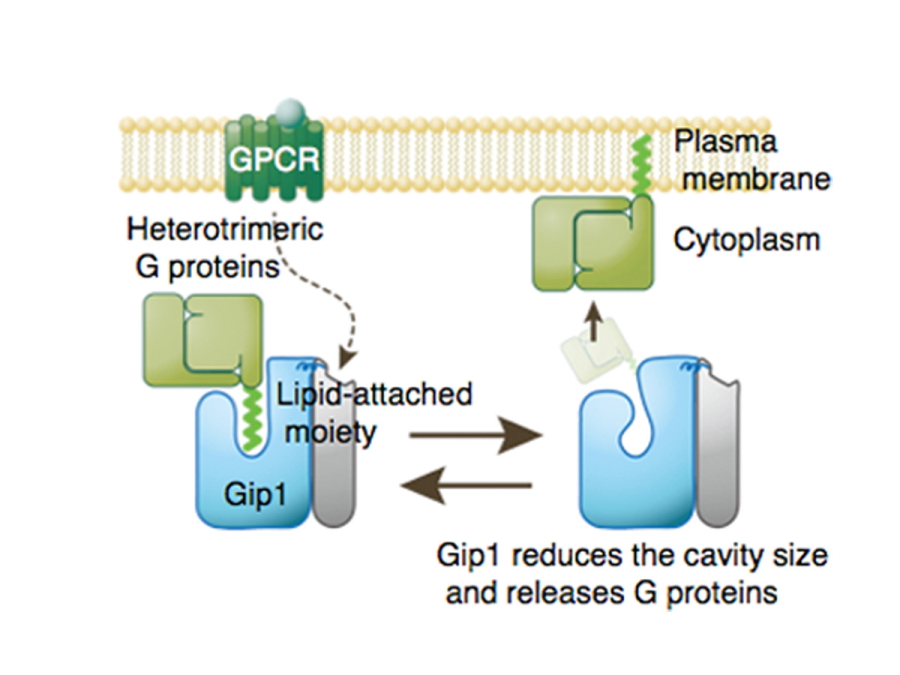
A schematic showing Gip1 sequestration of G proteins in the cytosol.
G proteins typically attach to the surface of the plasma membrane by anchoring their hydrophobic lipid-attached region into the membrane. However, Gip1 interacts with and masks this lipid-attached region of G proteins, so that G proteins are sequestered in the cytosol. Upon receiving signals from GPCR proteins, the size of the Gip1 binding cavity is reduced, which allows release of G proteins.
| Authors | Miyagawa T (1, 2), Koteishi H (2, 3), Kamimura Y (2), Miyanaga Y (1), Takeshita K (5, 6), Nakagawa A (5), Ueda M (1, 2)
|
|---|---|
| PubMed | 30401901 |
