Biomolecular Networks Laboratories
RNA Biofunction Laboratory
 Prof. HIROSE Tetsuro
Prof. HIROSE Tetsuro
Keywords:
Non-coding RNA (ncRNA), Phase separation, Membraneless organelle, Stress response, Disease
We study the architectural function of non-coding RNAs to build intracellular structures.
Transcriptome analysis has revealed that large portions of eukaryotic genomes produce numerous non-coding RNAs (ncRNAs), which expectedly play important regulatory roles in various biological processes. Our goal is to elucidate the functions of these ncRNAs and reveal the underlying new genetic code, thereby redefining the basic concept of genome function. We recently found that ncRNAs play architectural roles in membraneless organelles. Now we are studying the mode of action and cellular function of these ncRNAs using basic molecular and cellular experimental techniques combined with biophysical and bioinformatic tools.
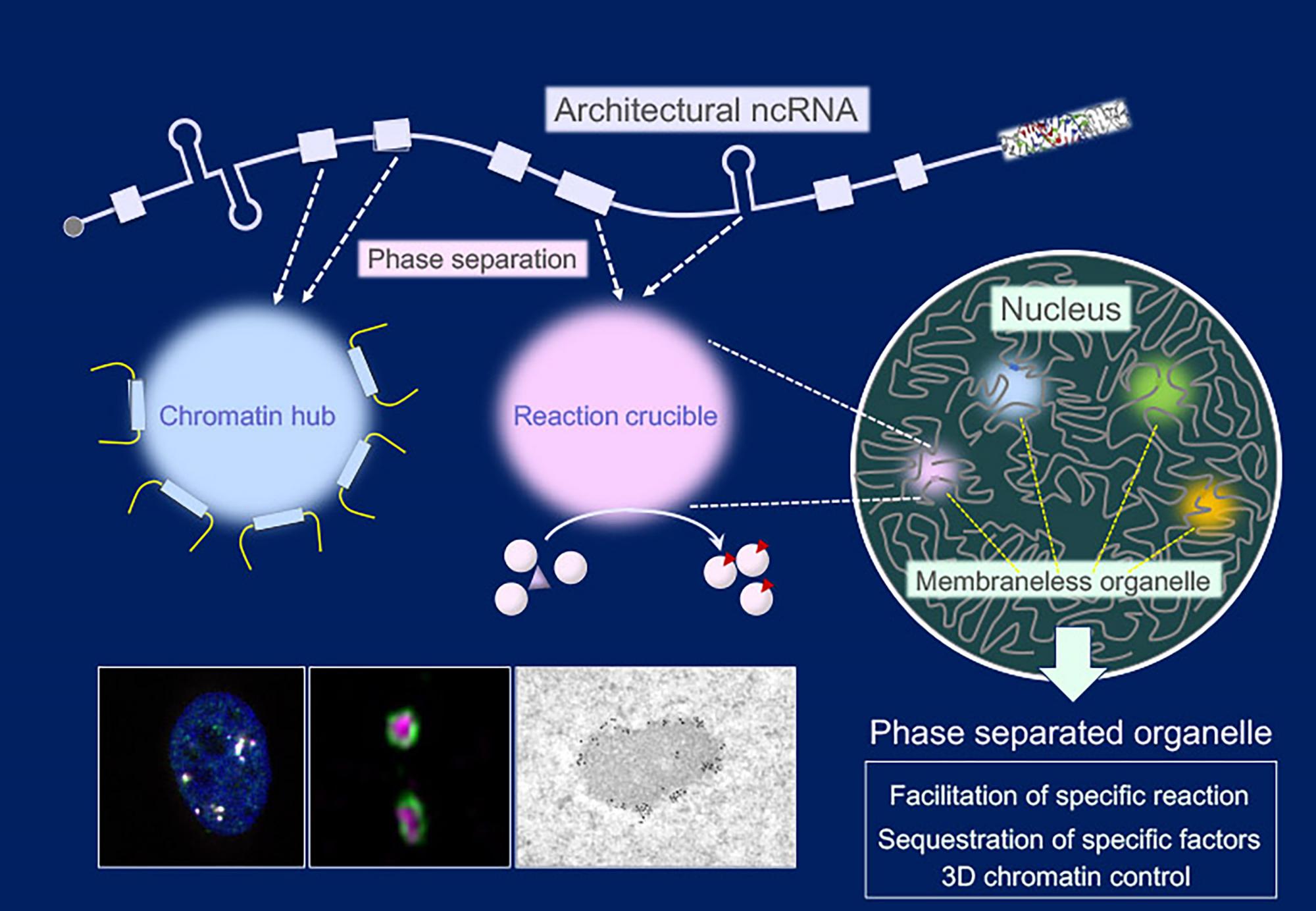
Architectural ncRNA can construct membraneless organelles through phase separation. The ncRNAs harbor distinct functional domains to function as chromatin hubs and reaction crucibles for specific biochemical reactions, particularly in the nucleus.
Images: Paraspeckle as an ncRNA-dependent membraneless organelle detected by a confocal microscope (left), a super-resolution microscope (middle) and an electron microscope (right).
Members
| Tetsuro Hirose (Professor) | hirose.tetsuro.fbs[at]osaka-u.ac.jp |
|---|---|
| Tomohiro Yamazaki (Associate Professor) | tyamazaki[at]fbs.osaka-u.ac.jp |
| Kensuke Ninomiya (Associate Professor) | ninomiya.kensuke.fbs[at]osaka-u.ac.jp |
| Ichiro Taniguchi (Assistant Professor) | taniguchi.ichiro.fbs[at]osaka-u.ac.jp |
| Kanako Kita (Assistant Professor, Graduate School of Medicine) | |
| Naoko Fujiwara (Assistant Professor) | nfujiwara.fbs[at]osaka-u.ac.jp |
| Rieko Takahashi (Technical Assistant) | |
| Hisako Asada (Technical Staff) | |
| Kanako Ezoe (Secretary) | |
| Hiroko Yasui (Secretary) | |
You could probably reach more information of individual researchers by Research Map and researcher's search of Osaka-U.
- ※Change [at] to @
Q&A
- What is your hot research topic?
- Decoding the ncRNA code
- Mechanism of phase separation or transition induced by ncRNA
- Mechanism of ncRNA bodies-chromatin intereactions
- Function of genomic repetitive elements
- Acquisition mechanism of species-specific traits
- What is your breakthrough or research progress in the last 5 years?
- Our group discovered a novel ncRNA function that works as a structural scaffold of membraneless organelles. The ncRNA can sequestrate multiple proteins with intrinsically disordered domains, thereby inducing liquid-liquid phase separation that is a driving force to construct the massive membraneless organelles. Multiple ncRNAs with the similar function were additionally identified as products of the human genome. Based on these results, we proposed to term these ncRNAs “architectural ncRNAs” as a new functional subcategory in ncRNAs.
- What kind of background do your lab members have?
- Our group members with various backgrounds join the laboratory after graduation of Faculty of Science, Engineering, Agriculture or Medicine.
- Do you collaborate with other institutions and universities?
- Our group has been collaborating with a number of laboratories in Japan (Hokkaido Univ., Univ. of Tokyo, AIST and RIKEN) as well as in foreign countries (CNRS in FR, MPI in DEU, Fraincis Crick Inst in UK, Univ. of Rochester in USA, Univ. of Western Australia in AUS and KAUST in SA)
- What kind of careers do your Lab's alumni go on to?
- Many of the former laboratory members successfully got the academic positions (e.g. Kumamoto Univ., Ritsumeikan Univ., NCGH and Univ. of Arizona) and the researcher position in bio-venture company.
- How do you develop your research?
- Understanding ncRNA functions and decoding the ncRNA code will reconstruct the basic concept on the mode of the genome functions and also facilitate the research by new perspective to understand the mechanism of various physiological events and diseases.
- Recent Representative Publications
- Hirose et al. (2022) Nat Rev Mol Cell Biol.
Ninomiya et al. (2021) EMBO J. 40, e107976
Yamazaki et al. (2021) EMBO J. 40, e107270
Ninomiya et al. (2020) EMBO J. 39, e102729
Yamazaki et al. (2018) Mol. Cell 70, 1038-1053
Chujo et al. (2017) EMBO J. 30, 1447-1462
Mannen et al. (2016) J. Cell Biol. 214, 45-59
Kawaguchi et al. (2015) PNAS 112, 4304-4309
Research Highlights
Publications (Research Articles, Reviews, Books)
2023
Functional domains of nuclear long noncoding RNAs: Insights into gene regulation and intracellular architecture
Curr Opin Cell Biol. 5:102250 2023 (PMID:37806294 DOI:10.1016/j.ceb.2023.102250.)
Nascent ribosomal RNA act as surfactant that suppresses growth of fibrillar centers in nucleolus.
Commun Biol 6(1):1129 2023 (PMID:37935838 DOI:10.1038/s42003-023-05519-1)
Shell protein composition specified by the lncRNA NEAT1 domains dictates the formation of paraspeckles as distinct membraneless organelles
Nat. Cell Biol. 25(11):1664-1675 2023 (PMID:37932453 DOI:10.1038/s41556-023-01254-1)
Satellite RNAs: emerging players in subnuclear architecture and gene regulation
EMBO J. 42(18):e114331 2023 (PMID:37526230 DOI:10.15252/embj.2023114331)
Nondomain biopolymers: Flexible molecular strategies to acquire biological functions
Genes Cells. 28(8):539-552 2023 (PMID:37249032 DOI:10.1111/gtc.13050)
Landscape of semi-extractable RNAs across five human cell lines
Nucleic Acids Res. 51(15):7820-7831 2023 (PMID:37463833 DOI:10.1093/nar/gkad567)
A guide to membraneless organelles and their various roles in gene regulation
Nat Rev Mol Cell Biol. 24(4):288-304 2023 (PMID:36424481 DOI:10.1038/s41580-022-00558-8)
Remarkable improvement in detection of readthrough downstream-of-gene transcripts by semi-extractable RNA-sequencing
RNA. 29(2):170-177 2023 (PMID:36384963 DOI:10.1261/rna.079469.122)
Long non-coding RNAs: definitions, functions, challenges and recommendations
Nat Rev Mol Cell Biol. 24(6):430-447 2023 (PMID:36596869 DOI:10.1038/s41580-022-00566-8)
2022
tRNA-like Transcripts from the NEAT1-MALAT1 Genomic Region Critically Influence Human Innate Immunity and Macrophage Functions
Cells. 11(24):3970 2022 (PMID:36552736 DOI:10.3390/cells11243970)
Triblock copolymer micelle model of spherical paraspeckles
Front Mol Biosci. 9:925058 2022 (PMID:36072433 DOI:10.3389/fmolb.2022.925058)
Micellization: A new principle in the formation of biomolecular condensates
Front Mol Biosci. 9:974772 2022 (PMID:36106018 DOI:10.3389/fmolb.2022.974772)
Statistical Thermodynamics Approach for Intracellular Phase Separation
Methods Mol Biol. 2509:361-393 2022 (PMID:35796975 DOI:10.1007/978-1-0716-2380-0_22)
lncRNA Neat1 regulates neuronal dysfunction post-sepsis via stabilization of hemoglobin subunit beta
Mol Ther. 30(7):2618-2632 2022 (PMID:35331906 DOI:10.1016/j.ymthe.2022.03.011)
2021
NEAT1 is essential for metabolic changes that promote breast cancer growth and metastasis
Cell Metab. 33(12):2380-2397 2021 (PMID:34879239 DOI:10.1016/j.cmet.2021.11.011)
Noncoding RNAs: biology and applications-a Keystone Symposia report
Ann N Y Acad Sci 1506(1):118-141 2021 (PMID:34791665 DOI:10.1111/nyas.14713)
Distinct RNA polymerase transcripts direct the assembly of phase-separated DBC1 nuclear bodies in different cell lines
Mol Biol Cell. 32(21):ar33 2021 (PMID:34495685 DOI:10.1091/mbc.e21-02-0081)
SPF45/RBM17-dependent, but not U2AF-dependent, splicing in a distinct subset of human short introns
Nat Commun. 12(1):4910 2021 (PMID:34389706 DOI:10.1038/s41467-021-24879-y)
Control of condensates dictates nucleolar architecture
Science. 373(6554):486-487 2021 (PMID:34326220 DOI:10.1126/science.abj8350)
m6A modification of HSATIII lncRNAs regulates temperature-dependent splicing
EMBO J. 40(15) e107976 2021 (PMID:34184765 DOI:10.15252/embj.2021107976)
ArcRNAs and the formation of nuclear bodies
Mamm genome. 33(2):382-401 2021 (PMID:34085114 DOI:10.1007/s00335-021-09881-5)
Paraspeckles are constructed as block copolymer micelles
EMBO J. 40(12) e107270 2021 (PMID:33885174 DOI:10.15252/embj.2020107270)
Small molecule targeting r(UGGAA)n disrupts RNA foci and alleviates disease phenotype in Drosophila model
Nat Commun. 12(1):236-236 2021 (PMID:33431896 DOI:10.1038/s41467-020-20487-4)
CRISPR-Mediated Mutagenesis of Long Noncoding RNAs
Methods Mol Biol. 2254:283-303 2021 (PMID:33326083 DOI:10.1007/978-1-0716-1158-6_18)
2020
Inhibition of the long non-coding RNA NEAT1 protects cardiomyocytes from hypoxia in vitro via decreased pri-miRNA processing
Cell Death Dis. 11(8):677 2020 (PMID:32826883 DOI:10.1038/s41419-020-02854-7)
Functional annotation of human long noncoding RNAs via molecular phenotyping
Genome Res. 30(7):1060-1072 2020 (PMID:32718982 DOI:10.1101/gr.254219.119)
NONO Is a Negative Regulator of SOX2 Promoter
Cancer Genomics Proteomics. 17(4):359-367 2020 (PMID:32576581 DOI:10.21873/cgp.20195)
Long non-coding RNA Neat1 regulates adaptive behavioural response to stress in mice
Transl Psychiatry 10(1):171-171 2020 (PMID:32467583 DOI:10.1038/s41398-020-0854-2)
Phase separation driven by production of architectural RNA transcripts
Soft Matter. 16(19):4692-4698 2020 (PMID:32396591 DOI:10.1039/c9sm02458a)
Detailed analyses of the crucial functions of Zn transporter proteins in alkaline phosphatase activation
Journal of Biological Chamistry 295(17):5669-5684 2020 (PMID:32179649 DOI:10.1074/jbc.RA120.012610)
Our ideal candidate (as a graduate student)
We are looking for a highly motivated person to work on our research topics as our lab member. Our lab welcomes the person who loves taking care of creatures, hand working and handcraft too. Any kind of background (such as your expertise or major) is available.
Contact
RNA Biofunction Laboratory, Graduate School of Frontier Biosciences, Osaka University,
1-3 Yamadaoka, Suita, Osaka 565-0871 Japan.
TEL: +81-6-6879-4675
E-mail: hirose.tetsuro.fbs[at]osaka-u.ac.jp (Prof. Tetsuro Hirose)
- ※Change [at] to @
