Biomolecular Networks Laboratories
Laboratory of Mitochondrial Dynamics
 Assoc. Prof. OKAMOTO Koji
Assoc. Prof. OKAMOTO Koji
Keywords:
Yeast, Mitochondria, Organelle, Mitophagy
Unraveling mitochondrial quality and quantity control mechanisms
Mitochondria are energy-converting organelles that act as "the power plants of the cell", and change their quantity in response to cellular energy demand. They also accumulate oxidative stress from reactive oxygen species generated during electron transport, and damaged mitochondria are selectively eliminated from the cytoplasm. These quality and quantity control systems involve mitochondria-specific autophagy (mitophagy), and numerous studies suggest that defects in these systems are associated with various human diseases. Mitophagy is a catabolic process conserved from yeast to humans that sequesters and degrades mitochondria as whole organelles. The aim of our study is to uncover the general principle of mitophagy at the molecular and cellular levels, and elucidate the physiological function of this degradation process as an intracellular quality and quantity control system.
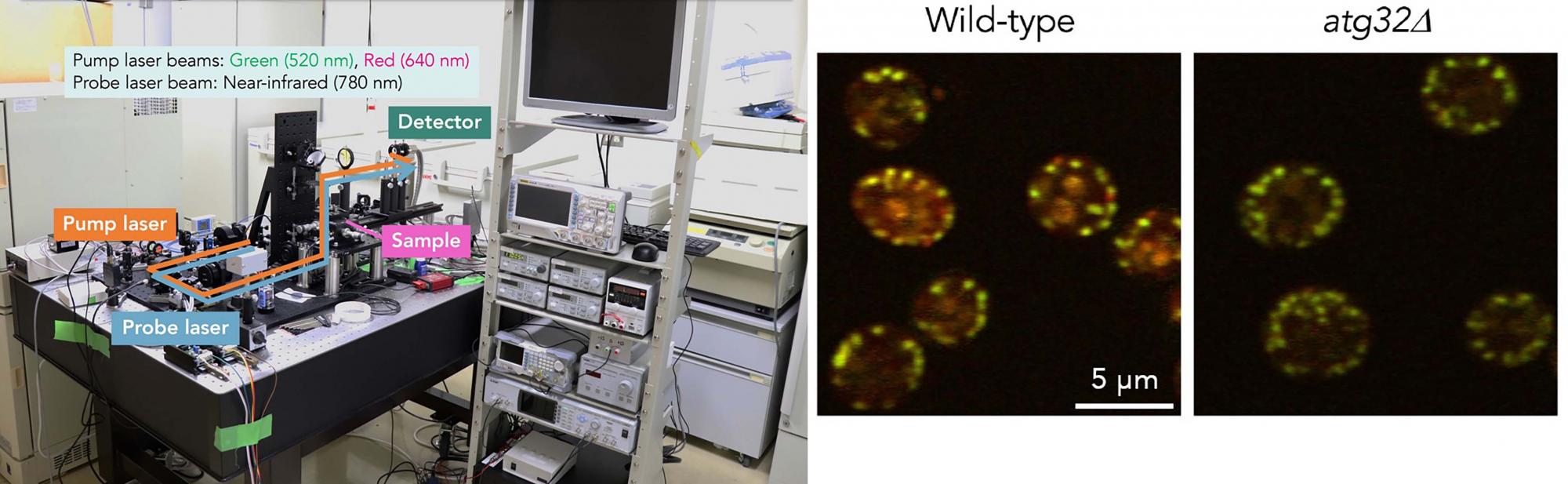
Custom-built photothermal microscopy (PTM)(left panel): Chromophore in mitochondria absorbs the pump laser energy and generates heat (only a few K). The refractive index around chromophore changes with heat. Variation in the refractive index induces deflection of the probe laser. Finally, the magnitude of probe laser deflection can be converted to optical signals, thereby visualizing mitochondria.
Label-free photothermal imaging of budding yeast (right panel): Using PTM, signals derived from intact mitochondria and vacuoles (lysosomal organelles) containing mitochondria delivered via mitophagy are seen in wild-type cells. The latter signal cannot be detected in cells lacking Atg32, a protein essential for mitophagy (atg32∆)
Members
| Koji Okamoto (Associate Professor) | okamoto.koji.fbs[at]osaka-u.ac.jp | ||
|---|---|---|---|
| Duan Lan(D5) | u256902b[at]ecs.osaka-u.ac.jp | ||
| Mitsutaka Kubota(D5) | u133374K[at]ecs.osaka-u.ac.jp | ||
| Yuki Nakayama(D4) | u343383c[at]ecs.osaka-u.ac.jp | ||
| Sayaka Nagano(D2) | u287877f[at]ecs.osaka-u.ac.jp | ||
| Peijin Li(D1) | u447747c[at]ecs.osaka-u.ac.jp | ||
| Rio Sato(D1) | u186686d[at]ecs.osaka-u.ac.jp | ||
| Qiao Liu(research student) | u060595i[at]ecs.osaka-u.ac.jp | ||
| Yuko Imada (Secretary) | imada-yuu[at]office.osaka-u.ac.jp |
You could probably reach more information of individual researchers by Research Map and researcher's search of Osaka-U.
- ※Change [at] to @
Q&A
- What is your hot research topic?
- Previous studies suggest that mitochondria are established independently from other organelles. However, it has recently become evident that they cooperate physically and functionally with other organelles such as the endoplasmic reticulum (ER), peroxisome, and lysosome. We found that selective degradation of mitochondria is also regulated via ER-localized proteins, and are now analyzing its molecular mechanisms.
- What is your breakthrough or research progress in the last 5 years?
- We have revealed that selective degradation of mitochondria is intimately linked to diverse processes including phospholipid methylation, protein N-terminal acetylation, phospholipid dephospholylation, nutrient/stress signaling, ER-associated degradation, and ER membrane protein insertion.
- What kind of background do your lab members have?
- The lab members have expertise in molecular biology, biochemistry, genetics, and cell biology using bacteria, yeast, and mammalian cultured cells as model systems.
- Do you collaborate with other institutions and universities?
- We are currently collaborating with researchers from Wakayama University, Nagoya City University, Johns Hopkins University (Baltimore, MA, USA), and the University of Groningen (Groningen, Netherlands).
- What kind of careers do your Lab's alumni go on to?
- Nine former lab members are now working at companies, two are taking a postdoc in Germany, and another is studying at a graduate school in the USA.
- How do you develop your research?
- We decipher basic principles underlying selective degradation of mitochondria at the molecular level in depth. Recently, using yeast, we have established a label-free method to detect selective degradation of mitochondria. We aim to elucidate the universality and diversity of selective mitochondrial degradation by making this method available to a variety of organisms other than model organisms used in the laboratory.
Research Highlights
Publications (Research Articles, Reviews, Books)
2024
Fluorescence Microscopy and Immunoblotting for Mitophagy in Budding Yeast
Methods in Molecular Biology (2845) 1-14 2024 (PMID:39115653 DOI:10.1007/978-1-0716-4067-8_1)
The nascent polypeptide-associated complex subunit Egd1 is required for efficient selective mitochondrial degradation in budding yeast
Scientific Reports 14(1):546. 2024 (PMID:38177147 DOI:10.1038/s41598-023-50245-7.)
2023
A ubiquitin-proteasome pathway degrades the inner nuclear membrane protein Bqt4 to maintain nuclear membrane homeostasis
Journal of Cell Science 136(19):jcs260930. 2023 (PMID:37694715 DOI:10.1242/jcs.260930)
The GET pathway serves to activate Atg32-mediated mitophagy by ER targeting of the Ppg1-Far complex
Life Sci Alliance 6( 4) e202201640 2023 (PMID:36697253 DOI:10.26508/lsa.202201640)
2022
An overview of the molecular mechanisms of mitophagy in yeast
Biochimica et Biophysica Acta - General Subjects 1866(11):130203. 2022 (PMID:35842014 DOI:10.1016/j.bbagen.2022.130203.)
2021
The protein N-terminal acetyltransferase A complex contributes to yeast mitophagy via promoting expression and phosphorylation of Atg32
J. Biochem. 170(2):175-182 2021 (PMID:34115119 DOI:10.1093/jb/mvab068)
Mitochondrial dynamics and degradation in the oleaginous yeast Lipomyces starkeyi
Genes Cells 26(8):627-635 2021 (PMID:34085353 DOI:10.1111/gtc.12875)
2020
Detection of mitophagy in mammalian cells, mice, and yeast.
Methods Cell Biol. 155: 557-579 2020 (PMID:32183977 DOI:10.1016/bs.mcb.2019.10.006)
2019
Repression of mitochondrial metabolism for cytosolic pyruvate-derived chemical production in Saccharomyces cerevisiae.
Microb. Cell. Fact. 0.872916667 2019 (PMID:31615527 DOI:10.1186/s12934-019-1226-6)
The Paf1 complex transcriptionally regulates the mitochondrial-anchored protein Atg32 leading to activation of_mitophagy.
Autophagy 1月14日 2019 (PMID:31525119 DOI:10.1080/15548627.2019.1668228)
A Mammalian Mitophagy Receptor, Bcl2-L-13, Recruits the ULK1 Complex to Induce Mitophagy.
Cell Reports 26: 338-345.e6 2019 (PMID:30625316 DOI:10.1016/j.celrep.2018.12.050)
2018
Mitochondrial Stasis Reveals p62-Mediated Ubiquitination in Parkin-Independent Mitophagy and Mitigates Nonalcoholic Fatty Liver Disease
Cell Metab. 28: 588-604 2018 (PMID:30017357 DOI:10.1016/j.cmet.2018.06.014)
The TORC1 signaling pathway regulates respiration-induced mitophagy in yeast
Biochem. Biophys. Res. Commun. 502: 76-83 2018 (PMID:29787763 DOI:10.1016/j.bbrc.2018.05.123)
The ER membrane insertase Get1/2 is required for efficient mitophagy in yeast
Biochem. Biophys. Res. Commun. 503: 14-20 2018 (PMID:29673596 DOI:10.1016/j.bbrc.2018.04.114)
The Nem1-Spo7 protein phosphatase complex is required for efficient mitophagy in yeast
Biochem. Biophys. Res. Commun. 496: 51-57 2018 (PMID:29305265 DOI:10.1016/j.bbrc.2017.12.163)
2017
Phosphatidic Acid and Cardiolipin Coordinate Mitochondrial Dynamics
Trends Cell Biol. 28: 67-76 2017 (PMID:28911913 DOI:10.1016/j.tcb.2017.08.011)
Investigation of yeast mitophagy with fluorescence microscopy and Western blotting
Methods in Molecular Biology 1759: 71-83 2017 (PMID:28337707 DOI:10.1007/7651_2017_11)
Assays for mitophagy in yeast
Methods in Molecular Biology 1567:337-347. 2017 (PMID:28276028 DOI:10.1007/978-1-4939-6824-4_20)
Our ideal candidate (as a graduate student)
We are looking for a highly motivated person to work on our research topics as our lab member. Our lab welcomes the person who loves taking care of creatures, hand working and handcraft too. Any kind of background (such as your expertise or major) is available.
Contact
Laboratory of Mitochondrial Dynamics, Graduate School of Frontier Biosciences, Osaka University,
1-3 Yamadaoka, Suita, Osaka 565-0871 Japan.
TEL: +81-6-6879-7970
E-mail: kokamoto[at]fbs.osaka-u.ac.jp (Assoc. Prof. Koji Okamoto)
- ※Change [at] to @
