Research
The centromere/kinetochore plays a fundamental role in accurate chromosome segregation during mitosis and meiosis in eukaryotes (Figure). Its functions include sister chromatid adhesion and separation, microtubule attachment, chromosome movement, and mitotic checkpoint control. Although chromosome segregation errors cause genetic diseases including some cancers, the mechanism by which kinetochores interact with microtubules of the spindle apparatus during cell division is not fully understood.
To understand kinetochore assembly and function in higher vertebrate cells, we are taking multidisciplinary approaches including molecular genetics, cell biology, biochemistry, structural biology, and genome biology.
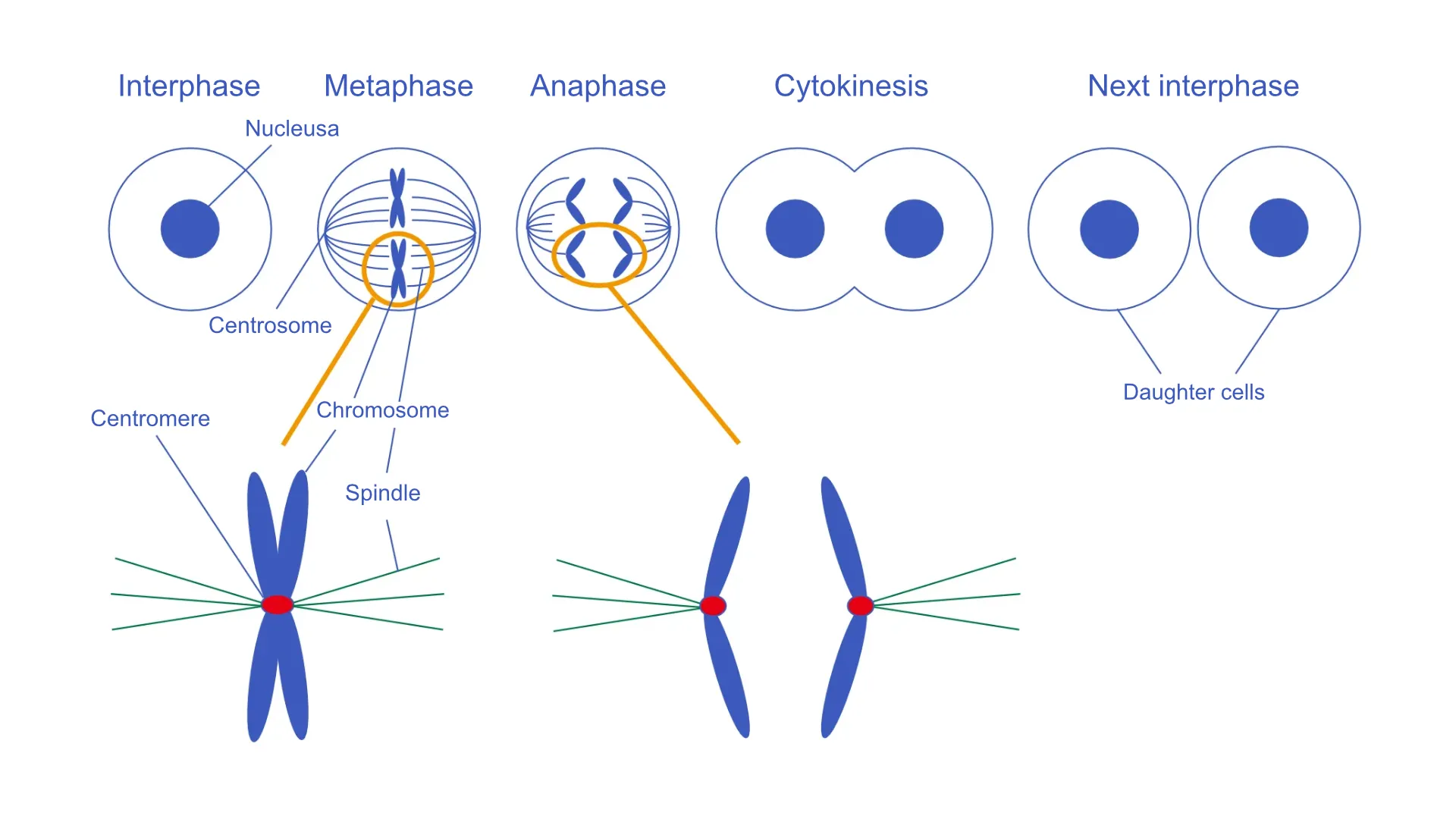
During mitosis, spindle microtubules bind to the centromere region on chromosomes, resulting in chromosome segregation.
We identified and characterized kinetochore proteins. We have also characterized centromeric DNA. The long-term goal of our lab is to understand how kinetochores are formed and how kinetochores function during mitosis. Our research will contribute to a better understanding of human health, as chromosomal abnormalities such as aneuploidy and translocations are strongly associated with tumor development and birth defects.
Currently, we are studying on
- Generation of mutant cells for kinetochore protein using cultured cells
- Functional analysis of kinetochore proteins using mutant cells
- Structural biology of the kinetochore protein complex
- Understanding centromere chromatin using genome biology
- Creation of artificial kinetochores using chromosome engineering
- Evolutionary conservation and diversity of Centromere/Kinetochores

01Generation of mutant cells for kinetochore protein using cultured cells
In the field of molecular biology, the most common method to analyze the function of a certain protein is to disrupt the function of the gene that encodes the protein and analyze its phenotype. We have long used DT40 cells derived from chicken B cells, which are suitable for generating genetically engineered cells because of their high frequency of homologous recombination when foreign genes are introduced into cells, and have generated many mutant cells for kinetochore proteins. In recent years, the use of genome editing technology has made the creation of mutant cells much more efficient. In addition to DT40 cells, we have also generated a wide variety of mutant cells in human and mouse cells.
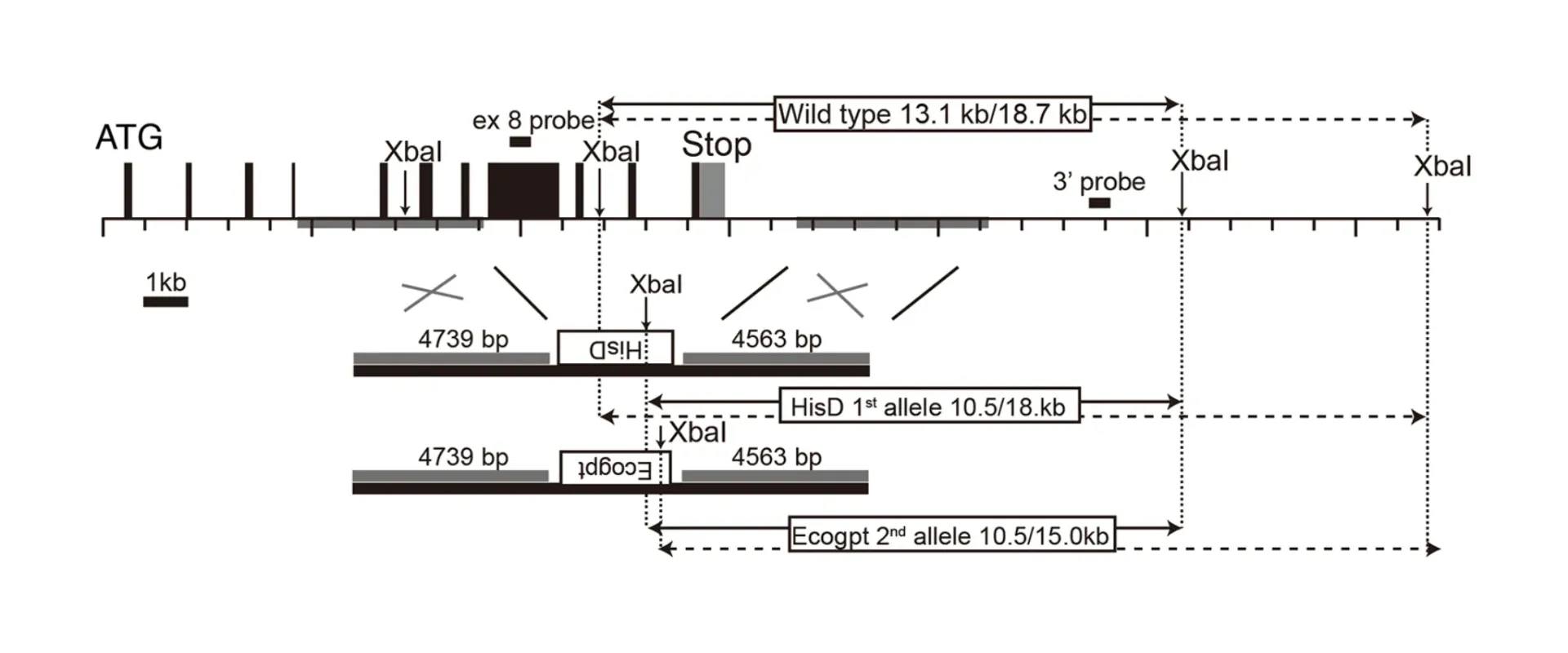
Using homologous recombination, we can freely modify an interesting genome locus.
02Functional analysis of kinetochore proteins using mutant cells
Once the various types of mutant cells have been created, their phenotypes are analyzed. Since we analyze the chromosome segregation, fluorescence microscopy is the most common method of analysis. Abnormal localization of kinetochore proteins and abnormal segregation of chromosomes will be observed under the microscope. In addition to analysis of fixed cells, we also incorporate techniques such as live cell imaging. In recent years, we have also used super-resolution microscopy with higher resolution to observe the fine structure of kinetochores.
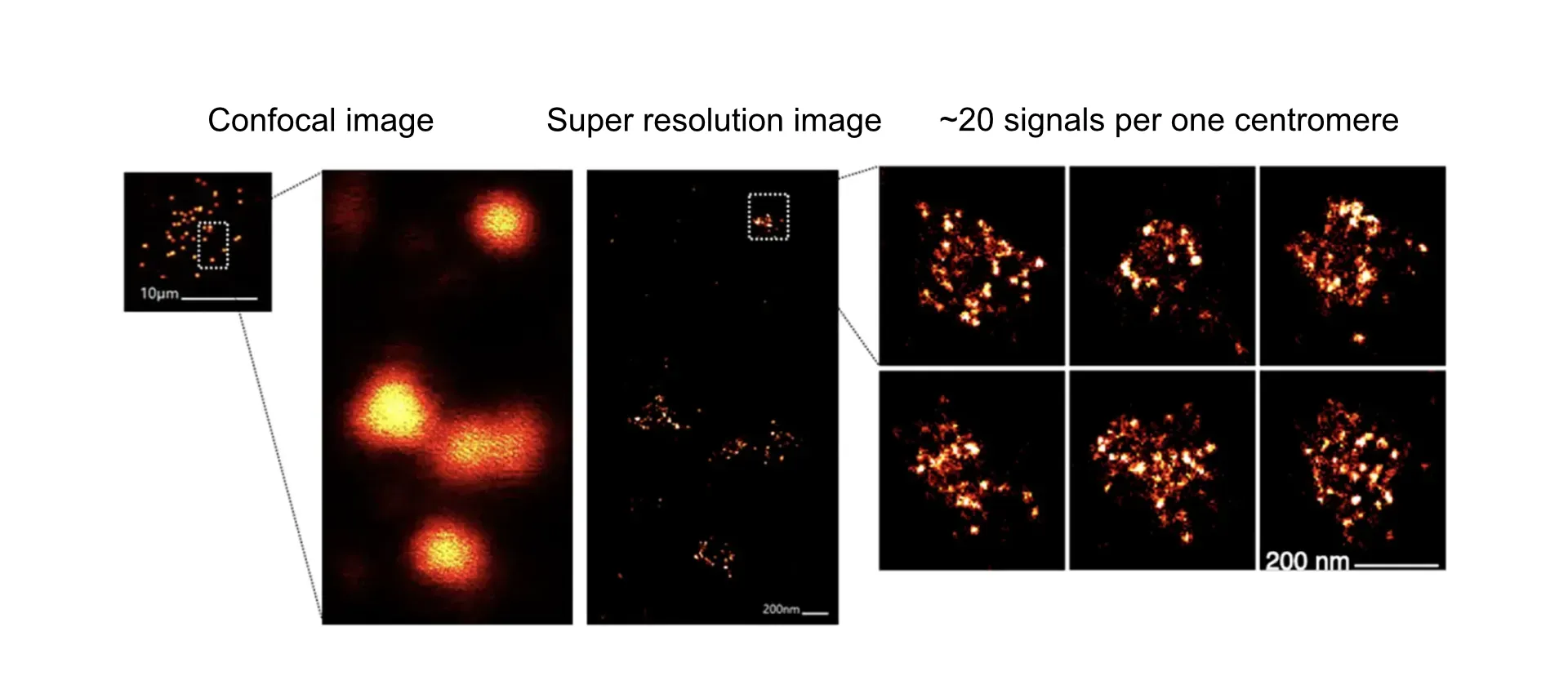
The kinetochore signal, which is visible at only one signal in a confocal microscope, is divided into ~20 signals.
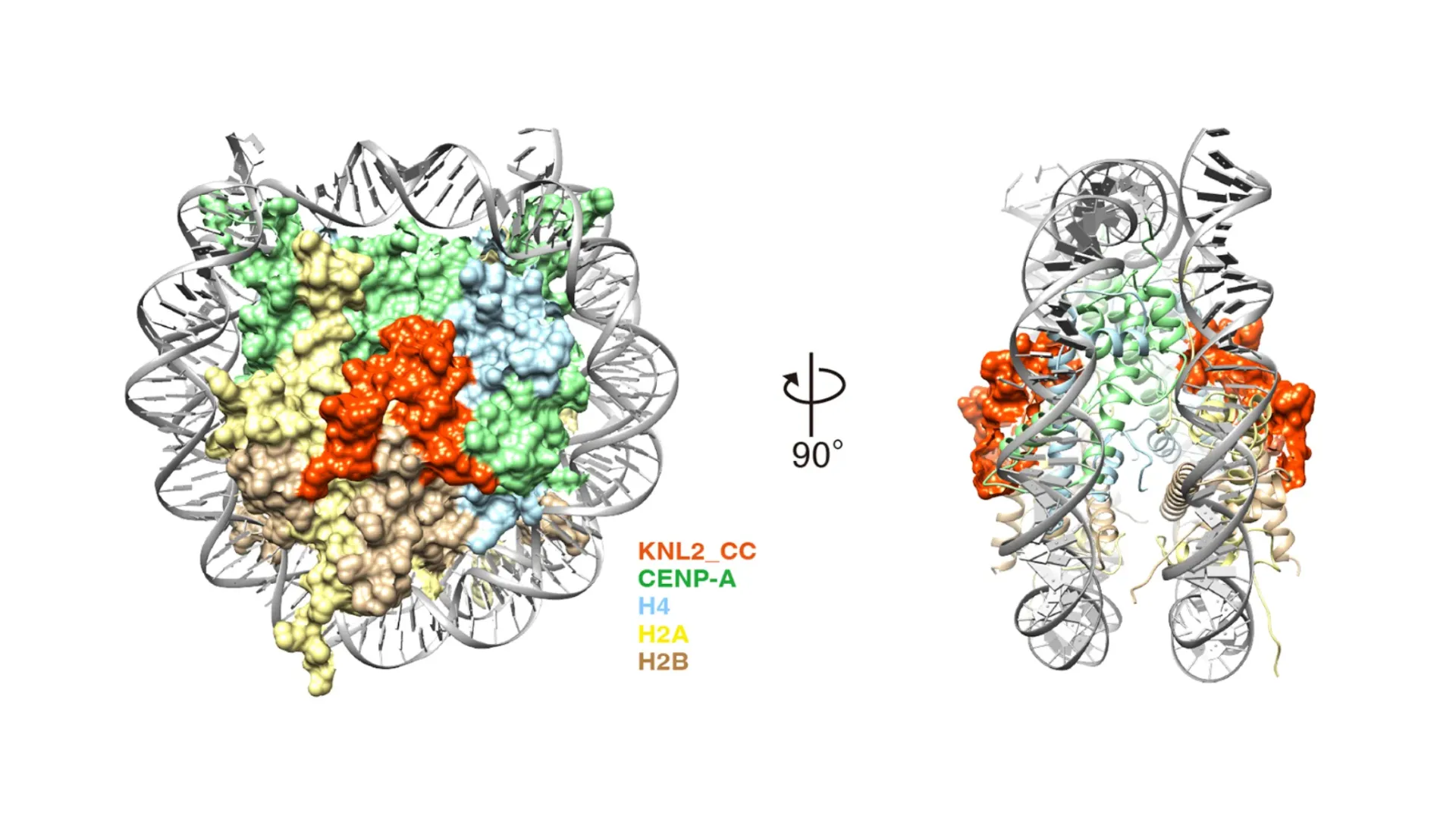
03Structural biology of the kinetochore protein complex
Many of the kinetochore proteins have been identified and are being actively studied to ask how the complex is formed. Structural biology methods are powerful for this purpose. In the past, complexes were reconstituted in vitro and then crystallized for X-ray structural analysis; now, if a reconstituted complex can be generated, its structure can be analyzed by cryo-electron microscopy. In addition to the analysis of reconstituted complexes, we are also challenging high-resolution analysis of kinetochore structures in cells.
04Understanding centromere chromatin using genome biology
Genomic DNA analysis in the centromere region has been delayed due to the presence of long repetitive sequences. Recently, however, with the advent of long-read sequencers, it has become possible to analyze long repetitive sequences. In this context, we also analyze centromere DNA and centromere chromatin. Since we have established an experimental system to generate centromeres without repetitive sequences in chicken DT40 cells, we are trying to use this advantage to characterize centromere chromatin using genomic approaches.
05Creation of artificial kinetochores using chromosome engineering
An experimental system for creating centromeres without repetitive sequences is one in which centromere regions are conditionally removed using chicken DT40 cells. By analyzing how centromeres are regenerated after centromere removal using such chromosome engineering techniques, we intend to elucidate the molecular mechanism by which centromere regions are specified.
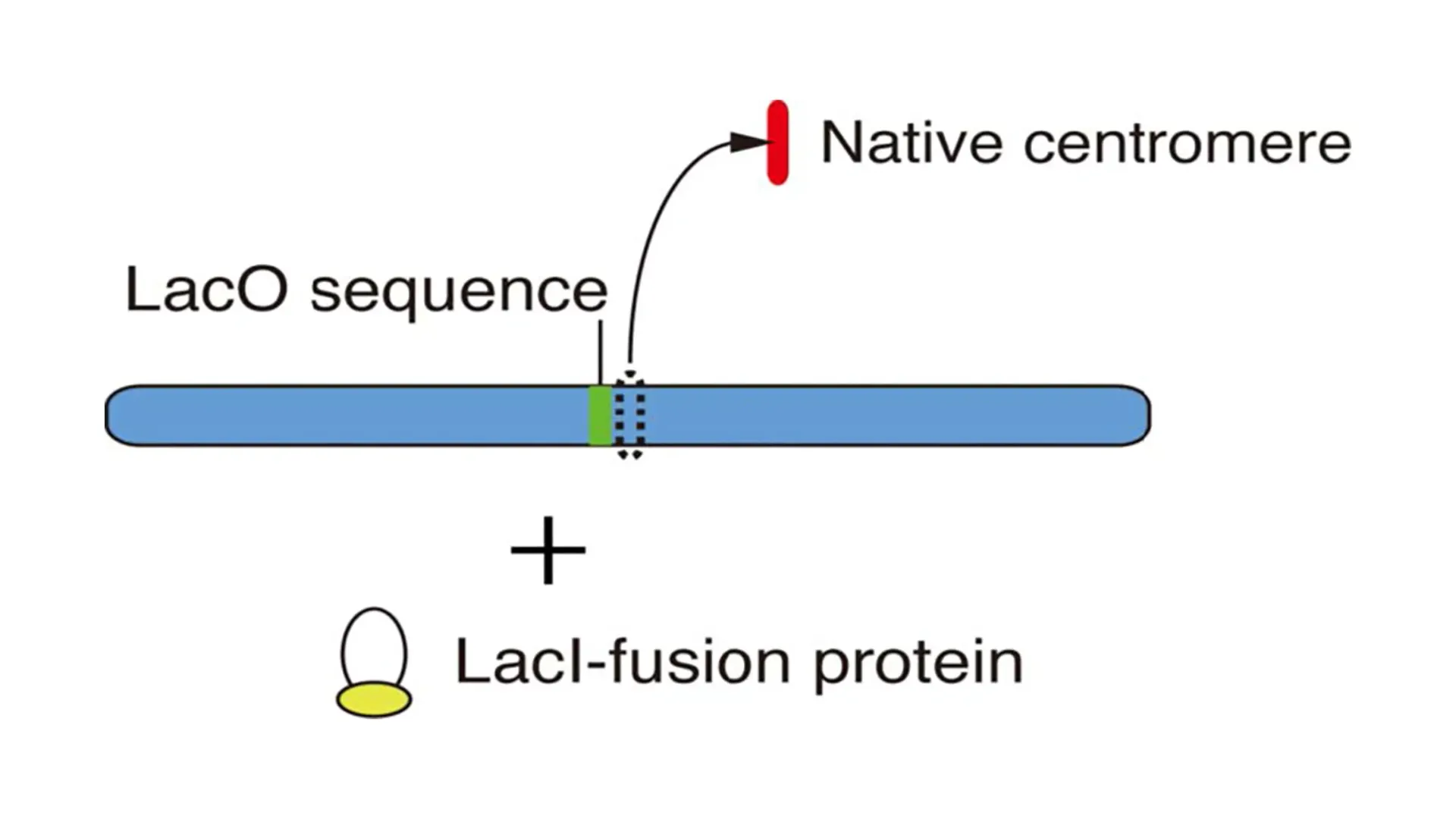
As shown in the figure, artificial kinetochores are created by ectopic localization of various proteins to non-centromere regions after removal of the native centromere. We are exploring the conditions under which we can efficiently construct artificial kinetochores not only in such experimental systems but also by using chromosome engineering techniques.
06Evolutionary conservation and diversity of Centromere/Kinetochore
Since the DNAs that constitute the centromere differ in sequence among organisms, kinetochore proteins are also not highly conserved among organisms through coevolution. In addition to this, the composition of kinetochores also differs among organisms. For example, some organisms lack the centromere-specific histone CENP-A, which defines the centromere in most organisms. How are centromeres specified in these organisms? We are also studying the conservation and diversity of centromere kinetochores in non-model organisms.

