Research Themes
Unveiling the secret of germline cells: the heir of the lives beyond birth and death of individuals
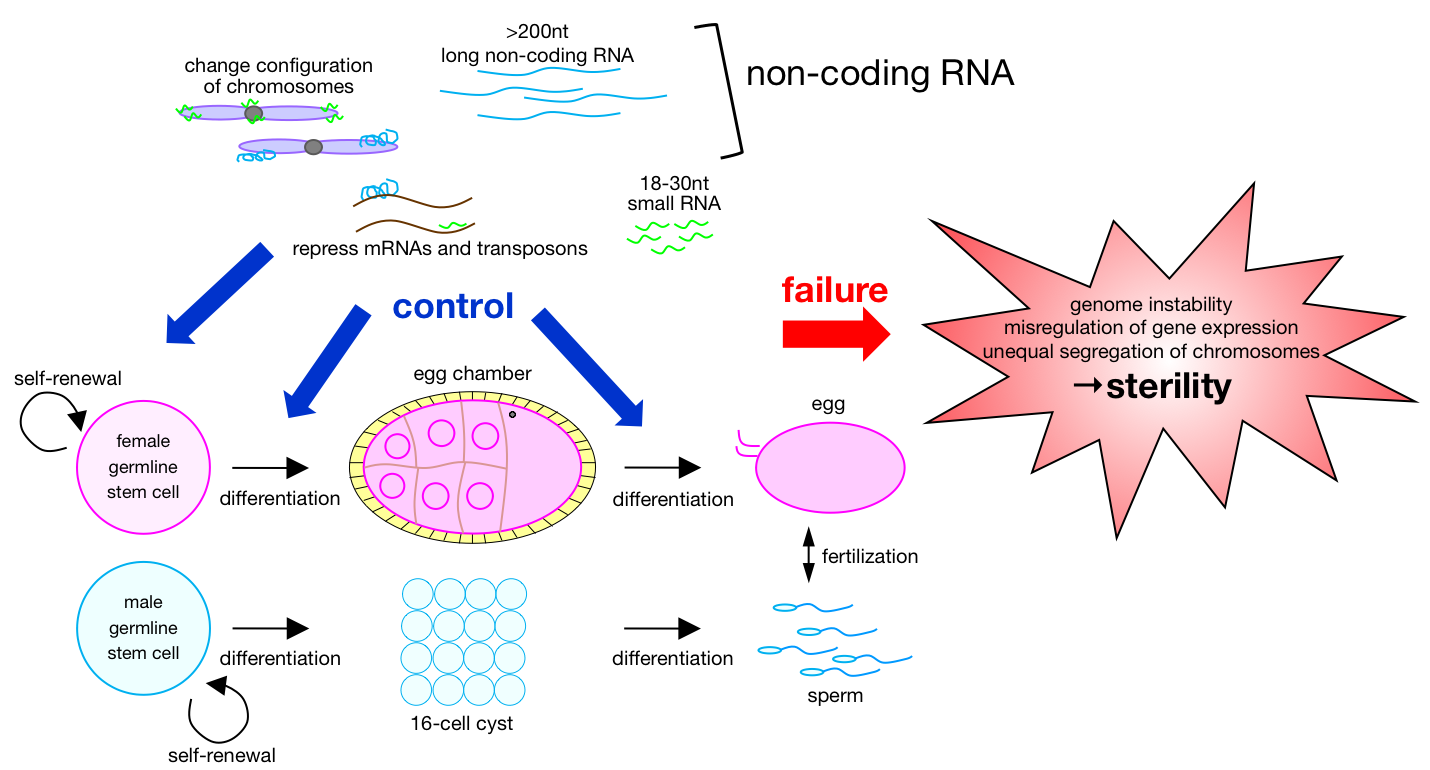
Thought higher animals do die as individuals, their species are maintained by sexual reproduction. Individuals are just like vehicles in which passenger –genetic information– can ride to be inherited to next generation by germline lineage. As such, germline cells are the most important cells to maintain the species. Drosophila melanogaster is one of the ideal model organisms we can study gametogenesis. How are germline stem cells maintained in the microenvironment, niche? What is the molecular mechanism that controls germline cells to mature into egg and sperm? How genome in germline cells safely guarded by non-coding RNA from transposons attacks? Our group has been addressing such molecular mechanisms for better understanding of gametogenesis.
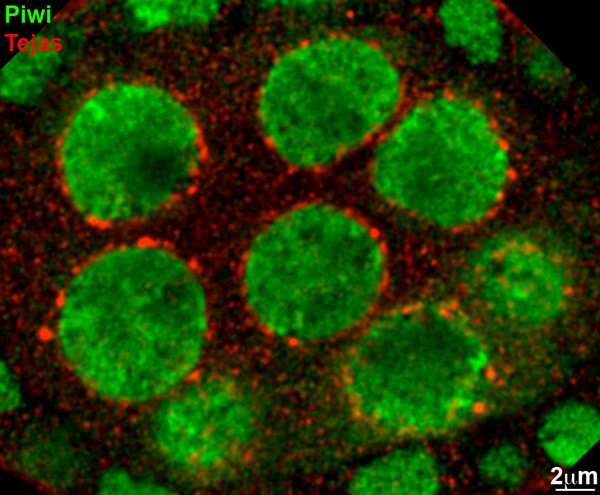
A confocal image showing one of Tudor domain proteins, Tejas (fly TDRD5 homolog), localized to nuage (red). Piwi involved in nuclear processing of piRNAs is shown in green. Tejas appears foci on the cytoplasmic face of the nurse cell nuclei.
Published Articles
Research articles
- *Kawaguchi S, *Xu X, Soga T, Yamaguchi K, Kawasaki R, Shimouchi R, Date S, Kai T. (2024) In silico screening by AlphaFold2 program revealed the potential binding partners of nuage-localizing proteins and piRNA-related proteins. eLife, 13:RP101967
*Co-first authors
doi: 10.7554/eLife.101967 - Xu F, Suyama R, Inada T, Kawaguchi S, Kai T. (2024) HemK2 functions for sufficient protein synthesis and RNA stability through eRF1 methylation during Drosophila oogenesis. Development, 151(14):dev202795
doi: 10.1242/dev.202795
PubMed ID: 40259744 - Suyama R, Cetraro N, Yew J Y, Kai T. (2023) Microbes control Drosophila germline stem cell increase and egg maturation through hormonal pathways. Commun. Biol., 6:Article number: 1287
doi: 10.1038/s42003-023-05660-x - Lin Y, Suyama R, Kawaguchi S, Iki T, Kai T. (2023) Tejas functions as a core component in nuage assembly and precursor processing in Drosophila piRNA biogenesis. J. Cell Biol., 222(10):e202303125
doi: 10.1083/jcb.202303125
PubMed ID: 37555815 - Iki T, Kawaguchi S, Kai T. (2023) miRNA/siRNA-directed pathway to produce noncoding piRNAs from endogenous protein-coding regions ensures Drosophila spermatogenesis. Science Advances, 9(29)
doi: 10.1126/sciadv.adh0397 - *Lim L-X, *Isshiki W, Iki T, Kawaguchi S, Kai T. (2022) The Tudor-domain containing protein, Kotsubu (CG9925), localizes to the nuage and functions in piRNA biogenesis in D. melanogaster. Frontiers in Molecular Biosciences (section RNA Networks and Biology), 9:Article 818302
*Co-first authors
doi: 10.3389/fmolb.2022.818302 - *Iki T, Takami M, *Kai T. (2020) Modulation of Ago2 Loading by Cyclophilin 40 Endows a Unique Repertoire of Functional miRNAs during Sperm Maturation in Drosophila. Cell Reports, 33(6):108380--108393
*Co-corresponding
doi: 10.1016/j.celrep.2020.108380 - Kawaguchi S, Ueki M, Kai T. (2020) Drosophila MARF1 ensures proper oocyte maturation by regulating nanos expression. PLoS One, 15(4):e0231114
doi: 10.1371/journal.pone.0231114
PubMed ID: 32243476 - Teo RYW, Anand A, Sridhar V, Okamura K, Kai T. (2018) Heterochromatin protein 1a functions for piRNA biogenesis predominantly from pericentric and telomeric regions in Drosophila. Nat. Commun., 9:1735
doi: 10.1038/s41467-018-03908-3 - Quénerch’du E, Anand A, Kai T. (2016) The piRNA pathway is developmentally regulated during spermatogenesis in Drosophila. RNA, 22(7):1044–1054
doi: 10.1261/rna.055996.116 - Patil VS, Anand A, Chakrabarti A, Kai T. (2014) The Tudor domain protein Tapas, a homolog of the vertebrate Tdrd7, functions in piRNA pathway to regulate retrotransposons in germline of Drosophila melanogaster. BMC Biol., 12:61
doi: 10.1186/s12915-014-0061-9
PubMed ID: 25287931 - Anand A, Kai T. (2014) Response to ‘Antisense piRNA amplification, but not piRNA production or nuage assembly, requires the Tudor-domain protein Qin’ (Correspondence). EMBO J., 33(6):540–541
doi: 10.1002/embj.201387548
PubMed ID: 24652837 - Lim RSM, Anand A, Nishimiya-Fujisawa C, Kobayashi S, Kai T. (2014) Analysis of Hydra PIWI proteins and piRNAs uncover early evolutionary origins of the piRNA pathway. Dev. Biol., 386(1):237–251
doi: 10.1016/j.ydbio.2013.12.007 - Pek JW, Ng BF, Kai T. (2012) Polo-mediated phosphorylation of Maelstrom regulates oocyte determination. Development, 139(24):4505–4513
doi: 10.1242/dev.082867 - Anand A, Kai T. (2012) The tudor domain protein Kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. EMBO J., 31(4):870–882
doi: 10.1038/emboj.2011.44 - Pek JW, Kai T. (2011) DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc. Natl. Acad. Sci. USA, 108(29):12007–12012
doi: 10.1073/pnas.1106245108 - Pek JW, Kai T. (2011) A role for Vasa in regulating mitotic chromosome condensation in Drosophila. Curr. Biol., 21(1):39–44
doi: 10.1016/j.cub.2010.11.051 - Patil VS, Kai T. (2010) Repression of retroelements in Drosophila germline via piRNA pathway by the tudor domain protein Tejas. Curr. Biol., 20(8):724–730
doi: 10.1016/j.cub.2010.02.046 - Pek JW, Lim AK, Kai T. (2009) Drosophila Maelstrom Ensures Proper Germline Stem Cell Lineage Differentiation by Repressing microRNA-7. Dev. Cell, 17(3):417–424
doi: 10.1016/j.devcel.2009.07.017 - Lim AK, Tao L, Kai T. (2009) piRNAs mediate post-transcriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J. Cell Biol., 186(3):333–342
doi: 10.1083/jcb.200904063 - Lim AK, Kai T. (2007) Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA, 104(16):6714–6719
doi: 10.1073/pnas.0701920104 - Kai T, Williams D, Spradling AC. (2005) The expression profile of purified Drosophila germline stem cells. Dev. Biol., 283(2):486–502
doi: 10.1016/j.ydbio.2005.04.018 - Kai T, Spradling A. (2004) Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature, 428(6982):564–569
doi: 10.1038/nature02436 - Kai T, Spradling A. (2003) An empty Drosophila stem cell niche reactivates the proliferation of ectopic cells. Proc. Natl. Acad. Sci. USA, 100(8):4633–4638
doi: 10.1073/pnas.0830856100
Review articles
- Suyama R, Kai T. (2024) piRNA processing within non-membrane structures is governed by constituent proteins and their functional motifs. FEBS J.,
doi: 10.1111/febs.17360 - Arakawa K, Hirose T, Inada T, Ito T, Kai T, Oyama M, Tomari Y, Yoda T, Nakagawa S. (2023) Nondomain biopolymers: Flexible molecular strategies to acquire biological functions. Genes to Cells,
doi: 10.1111/gtc.13050
PubMed ID: 37249032 - Niwa R, Kai T. (2020) Editorial overview: Stem cells orchestrate oogenesis: a lesson from the fruit fly, Drosophila melanogaster. Curr. Opin. Insect Sci., 37:iii–v
doi: 10.1016/j.cois.2020.03.001 - Gleason RJ, Anand A, *Kai T, *Chen X. (2017) Protecting and diversifying the germline. GENETICS, 208(2):435–471
(A part of FLY BOOK) *Co-corresponding
doi: 10.1534/genetics.117.300208 - Lim RSM, Kai T. (2015) A piece of the pi(e): the diverse roles of animal piRNAs and their PIWI partners. Sem. Cell Dev. Biol., 47–48:17–31
(Special Issue on Coding and Non-coding RNAs)
doi: 10.1016/j.semcdb.2015.10.025 - Pek JW, Anand A, Kai T. (2012) Tudor domain proteins in development (Invited Review Article). Development, 139:2255–2266
doi: 10.1242/dev.073304 - Pek JW, Patil VS, Kai T. (2012) The piRNA pathway and the potential processing site, the nuage, in the Drosophila germline. Dev. Growth Differ., 54(1):66–77
doi: 10.1111/j.1440-169X.2011.01316 - Pek JW, Kai T. (2011) Non-coding RNAs enter mitosis: functions, conservation and implications. Cell Div., 6:6
doi: 10.1186/1747-1028-6-6
PubMed ID: 21356070 - Pek JW, Kai T. (2009) Conserved germline organelle, nuage, serves as site for processing of Piwi-interacting RNAs. (in Japanese). Exp. Med., 27:393–399
- Kai T. (2004) Germline stem cells and their niches. (in Japanese). Tanpakushitsu Kakusan Koso, 49(6):710–717
- Spradling A, Drummond-Barbosa D, Kai T. (2001) Stem cells find their niche. Nature, 414(6859):98–104
doi: 10.1038/35102160
Protocol articles
- Lim RS, Osato M, Kai T. (2015) Isolation of undifferentiated female germline cells from adult Drosophila ovaries. Curr. Protoc. Stem Cell Biol., 34:2E.3.1–2E.3.14
doi: 10.1002/9780470151808.sc02e03s22