The origin of such various life functions as energy metabolism,
material metabolism, self-multiplication and environmental adaptability is
the flow of energy, signal and material through the complex networks formed
by macromolecules such as proteins and nucleic acids.
An enormous number of those macromolecules play each role just like
purposefully designed machines and maintain the complex network activities.
They could be called molecular machine, but they have characteristics quite
different from human-made ones in the following aspects:
(1) Designed with individual atoms as functional parts and therefore of nanometer size
(2) Self-organize the structure at the right time and place in the cellular organization
(3) Have flexibility and adaptability because of weak interactions of hydrogen bonding that forms the structure
The proton (H+) plays important roles in these functions and mechanisms,
in a similar way to what electrons do in semiconductor devices.
In the activities of living cells, where the water solution is
the basic medium for all the reactions, protons are the carriers of
the signal and energy.
Therefore, it is essential to clarify the mechanisms of macromolecular functions based on the behaviors of proton movements
in order to understand the activities of life at molecular level.
The aim of this project is to try to understand the physical processes
and mechanisms unique to biological molecular machines,
which efficiently transmit, amplify and transform extremely fine energy
and signal of conformational changes through proton exchange.
The research effort will be concentrated on
the atomic resolution structure analysis of macromolecular complexes
by X-ray diffraction and electron microscopy
and the analysis of energy transduction dynamics of the systems
by measuring their sub-nanoscale movements and proton flows
that are close to the present limit of experimental detection.
The results are expected to produce important clues to
the unknown principle of the macromolecular machinery.
The methods of structure and functional analyses of macromolecular complexes
developed by these studies will be applicable to
many other systems of important functions for better understandings
of the mechanisms and would make a great contribution to medical applications
as well.
These studies are also expected to give us some clues to the development
of a novel energy conversion technology
that is as efficient and well regulated as the biological system.
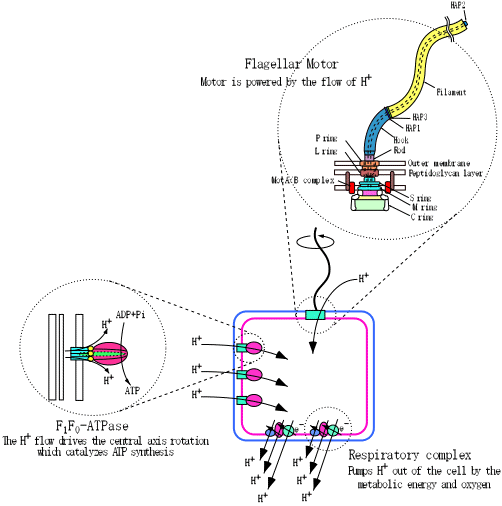
We chose the bacterial flagellum as the target system of our research project.
The bacterial flagellum is composed of a switchable rotary motor,
a universal joint and a helical propeller, all of
which are built by self-assembly of specifically designed component proteins.
The motor is powered by the proton motive force, just as F1F0ATPsynthase.
The component proteins are exported from the cytoplasmic side
to the distal end of the structure under construction
through its narrow central channel by the export apparatus
located at the base of the flagellum.
The assembly occurs at the distal end, where a capping complex helps
its efficient assembly process.
All of these functions are supported by mechanisms that are not yet known.
But, one thing is at least clear that we must see the atomic arrangements
and the dynamics of this huge molecular assembly to understand
how these intricate and dynamic functions are exhibited.
Groups of the project includes:
(1) NanoAssembly: will study the regulation mechanisms of macromolecular assemblies by X-ray crystallography and fiber diffraction.
(2) NanoSwitching: will approach the switching mechanisms of macromolecular assemblies by electron cryomicroscopy and image analysis.
(3) NanoMechanics: will look into the mechanisms of force generation and energy flow by optical and electrical measurements together with genetic engineering techniques.